Study contribution
Nowadays, Escherichia coli remains as one of main human pathogens contaminating foods. The consumption of beef and pork meet is one of the most important foods for human diet. In this study, two kind of meat samples were evaluated, and some E. coli isolates were detected containing stx2 and/or fliCH4 genes in beef and pork meat. These isolates were characterized as STEC non-O157:H7 being a potential risk for the consumers. Multiple antibiotic resistance was found in several isolates. This work demonstrates that the beef and pork meat can contains E. coli STEC non-O157:H7 representing a risk for the human health. This is the first report in Sonora, Mexico for STEC non-O157:H7.
Introduction
The ability of E. coli to cause disease in humans is associated with the production of Shiga toxins (stx1 and stx2).1 The Shiga toxin-producing E. coli (STEC) O157:H7 serogroup was first recognized in 1982 in the US when a massive outbreak of hemorrhagic colitis occurred due to the consumption of contaminated hamburgers,2,3 emerging as an important pathogen for humans.4 Beef and swine are asymptomatic carriers of this organism; however, beef is considered the main reservoir of virulent enterohemorrhagic strains.5 Once humans ingest this pathogen (infective dose is from 10 to 100 cells/g of food),6-8 it takes a 3- to 9-day incubation period before symptoms start to appear, and these symptoms include severe cramping (abdominal pain), diarrhea (initially liquid and later accompanied by bleeding), vomiting, and low or no fever, eventually reaching a stage of permanent renal function loss called Hemolytic Uremic Syndrome (HUS).9,10
The main routes of transmission of STEC to humans include from person to person through the fecal-oral route,1,6 the consumption of contaminated foods such as unpasteurized milk, water, fruits, vegetables, and raw beef, or through direct contact with animal feces.8,9 In the case of beef, contamination generally occurs during the slaughter of animals and carcass preparation.10
O157:H7 is the predominant serotype associated with outbreaks and cases of STEC infections worldwide.11 There are a growing number of non-O157 serogroups related to clinical symptoms such as mild to severe diarrhea, hemorrhagic colitis (HC) and even HUS.12,13 This E. coli non-O157 serogroup cluster has been termed the “big six”, and includes O26, O45, O103, O111, O121, and O145.14
The recognized genes to detect O157:H7 are stx1, stx2, eaeA, and rfB;15 while the “big six” serogroup show variations in the genetic composition of the wzx gene (O antigen flippase).16 The rfB gene is responsible for the biosynthesis of the O157 antigen, which is specific for detecting the O157:H7 serotype.12 The intimin protein, produced by the eaeA gene, is important for HUS development and is involved in the process of bacterial adherence to the epithelial cells of the human intestine (attaching and effacing).6,13
Several studies have demonstrated that the use of antibiotics to treat patients diagnosed with STEC and symptoms of diarrhea or bloody diarrhea may allow the development of HUS. Other reports showed the opposite, the alleviation of HUS symptomatology to milder symptoms.14,17,18 Besides, in vitro studies showed that sub-inhibitory antibiotic concentrations to treating O157:H7 STEC increase the amount of free Shiga toxin in the culture medium.18 Although antibiotic prophylaxis or therapy in STEC disease is still not the preferred treatment for patients with STEC, the knowledge on the antibiotic’s resistance/susceptibility of the native local STEC strains is always desired.
It is estimated that 112 752 cases of disease associated with this STEC non-O157 cluster occur each year, while O157:H7 is associated with 63 153 cases per year in countries such as Germany, Japan and the US, leading to 20 deaths each year and an annual economic loss of 255 million US dollars.19 A study performed in the United States over two years that included 4 133 ground beef samples identified 1 006 strains (24.3 %) of STEC carrying stx1 and/or stx2.20) There are few reports for Latin America. 274 half-beef sponges were processed in Argentina; E. coli O157 was isolated in four (n:1.4 %), of which two were characterized as stx2c(vh-a) eae/ehxa, and the other two as non-toxic.21 In Santiago, Chile, contamination by non-O157 was reported in 10 % of ground meat samples, with 56 isolates obtained from 43 positive samples showing a 41 % incidence of the stx2 gene (23/56) and a 10.7 % incidence (6/56) of the combination of the three pathogenicity genes (stx1, stx2, hlyA).22 In Colombia, the presence of E. coli O157:H7 was detected in 0.45 % (n:244) of bovine carcasses and for generic E. coli biotype 1 it was 0.89 %.23 In Mexico, the presence of E. coli O157:H7 was detected in 66.66 % (6/9) of beef and pork carcass samples in the Comarca Lagunera,24 while a 5 % incidence (2/40) was detected in beef cuts (rib eye sirloin, New York) in Monterrey, Nuevo León.25 Cabrera-Maldonado et al.26 analyzed 20 samples of ground beef from supermarkets in the city of Puebla and did not detect any O157:H7. In the northwest region of Mexico, where the State of Sonora is located, the incidence of E. coli was reported to be 38.24 % (13/34) in ground beef samples taken from Culiacan, Sinaloa, with no detection by real-time PCR of E. coli O157:H7 isolates among these samples.27
Mexico produces a high volume of meat (1.88 and 1.32 million tons of beef and pork, respectively) for both consumption and export (8 %-10 %).28 In the State of Sonora, meat production is an important economic activity, and this region has the second and tenth highest pork and beef and pork production, respectively, in Mexico. It is important to conduct studies to detect STEC O157 and non-O157 in this region to prevent disease outbreaks caused by these enteropathogenic bacteria. TIF slaughterhouses in Mexico, are animal slaughter facilities with mechanized processing permanently monitored for their sanitary conditions and regulated by international standards of quality and hygiene. In Mexico, the Secretaría de Agricultura y Desarrollo Rural (Sagarpa) and the standard NOM-194-SSA1-2004 establish the directives for slaughterhouses.29 Although TIF slaughterhouses offer quality meat and optimal sanitary conditions, the non-TIF slaughterhouses do not have mechanized systems and cannot comply with the international quality standards which results on poor quality meat management and suboptimal sanitary conditions.
The goals of this study were 1) to detect STEC O157 and non-O157 E. coli in meat products from TIF and non-TIF slaughterhouses, as well as butcheries in southern Sonora, Mexico; and 2) to verify the antibiotic resistance/susceptibility patterns of E. coli strains, which contain any toxin-producing gene. We hypothesized that 1) STEC O157:H7 is not present in meat samples in southern Sonora and 2) that the resulting non-O157 isolates will contain multiple resistance patterns to antibiotics.
Materials and methods
Ethical statement
No animal care and use Committee approval was obtained because meat samples were obtained from commercial area of slaughterhouses and in butcheries in the surrounding area to Obregon city in Sonora, Mexico.
Sample collection
Meat samples were collected from two types of slaughterhouses, TIF and non-TIF and from twelve butcheries in the urban area of Ciudad Obregon in Sonora, Mexico. Meat samples from the TIF slaughterhouse were taken from October through November 2015 and from the non-TIF slaughterhouse from October through December 2015 and from 12 local butcheries from March through December 2016, for 190 meat samples. The sampling details (periods, frequency of visits and specific comments on sampling) are provided in Table S1. Fifty-two pork samples were taken from a TIF slaughterhouse (19 loins, 19 ribs, and 14 pulps). A total of 55 beef samples (17 loins, 17 ribs, and 21 pulps) and 33 pork samples (10 loins, 10 ribs, and 13 pulps) were obtained from a non-TIF slaughterhouse. Finally, 50 ground beef samples were obtained from 12 local butcheries. Samples from the TIF slaughterhouses were taken from the freezing chamber (-25 to -30 oC) and the cutting room (4 to 8 oC); and at the non-FIT slaughterhouse samples were collected from the cold room (0 oC) and the cutting room (25 oC). The ground beef samples from the butcheries were collected from refrigerators kept at 4 oC from bulk ground meat containers.
We were unable to conduct a systematic sampling design, instead sampling in this study was transversal, which means that each cut type from the slaughterhouses was sampled at least three times during the sampling period. This type of sampling was performed since the slaughterhouses could not offer visits on a fixed day of the week, or the same type of cut for our weekly visits since this depended on meat availability and the demand for a certain cut type on that day. Nevertheless, we decided to obtain at least five samples of each cut type from the slaughterhouses (Table 1). In the local butcheries, we obtained three to five ground beef samples for ten butcheries, and in a few butcheries a single sample was taken either because they no longer carried out or they ran out of the product at the time of the visit.
Table 1 Number of presumptive O157 isolates obtained from meat samples
| Meat cuts | No. of presumptive isolates obtained/ No. samples | |||
| TIF | non-TIF | Commercial | ||
| Pork | CRP | 0/7 | 4/6 | - |
| Pork | CutRP | 0/7 | 17/7 | - |
| Pork | CRR | 1/9 | 4/5 | - |
| Pork | CutRR | 0/10 | 0/5 | - |
| Pork | CRL | 2/9 | 24/5 | - |
| Pork | CutRL | 2/10 | 33/5 | - |
| Beef | CRP | - | 5/10 | - |
| Beef | CutRP | - | 20/11 | - |
| Beef | CRR | - | 10/8 | - |
| Beef | CutRR | - | 23/9 | - |
| Beef | CRL | - | 23/8 | - |
| Beef | CutRL | - | 22/9 | - |
| Beef | *Ground beef | - | - | 34/50 |
| Total | Samples | 190 | ||
| Total | Isolates | 224 | ||
CRP= Cold room pulp; CutRP= Cutting room pulp; CRR= Cold room ribs; CutRR= Cutting room ribs; CRL= Cold room loin; CutRL= Cutting room loin.
* Ground beef from local butcheries.
Two hundred (slaughterhouses), or fifty (butcheries) grams were collected from each meat sample, placed in a plastic bag, hermetically sealed, and kept refrigerated in coolers at 4 to 8 oC and processed for further analysis in the following 24 hours.
Isolation of presumptive E. coli O157:H7 and non-O157
For isolating presumptive E. coli O157 and STEC non-O157 strains twenty-five grams of meat were maintained in 225 mL of Butterfield´s phosphate buffer and then shaken vigorously and incubated at 37 oC overnight (enrichment step). Fifty µL of 10-2 and 10-4 dilutions were spread in duplicate onto TC-SMAC agar and Rainbow® agar O157, and the plates incubated at 37 °C for 18-24 h for screening of typical E. coli colonies on TS-SMAC (colorless or neutral/gray with smoky center and 1-2 mm in diameter) and on Rainbow® agar O157 (black to blue-black colonies).30 Typical colonies were selected and purified by two to three successive passes on chromogenic agar; then, presumptive strains were kept in tryptic soy broth (TSB) (37 oC for 24h) and stored by triplicate in 20 % glycerol at -80 oC until used for biochemical and molecular analysis. The morphological and biochemical characterization were performed in accordance with the Mexican Official Standard.31 Isolates, which showed the biochemical profile for E. coli (see section below) and the coloration confirmed on Rainbow® agar O157 were used for molecular analyses.
Biochemical characterization of isolates
The morphological and biochemical characterization was performed in accordance with Appendix H of the Mexican Official Standard31 and included a Gram stain, the oxidase reaction (Merck KGaA, Cat. No. 113300, Darmstadt, Germany), the catalase test, and IMViC. This last test was used to identify enterobacteria; briefly, isolates were incubated in the following: 1) MIO medium (Difco Cat. No. 273520, Le Pont de Claix, France) to demonstrate the production of indole; 2) Methyl Red and Voges-Proskauer broth (Bioxon, Cat. No. 211691, Becton Dickinson de México S.A. de C.V, Cuautitlán Izcalli, México) to analyze the production of carboxylic acids (lactic, succinic, acetic, and formic) from glucose fermentation, as well as to detect the production of acetyl-methyl-carbinol from glucose fermentation to differentiate E. coli from the Klebsiella and Enterobacter genera; and 3) Simmons Citrate agar (Bioxon, Cat. No. 8245170) to determine the usage of citrate as a sole carbon source.32)
Extraction and quantification of bacterial DNA
Bacterial DNA from STEC O157 presumptive isolates was extracted with the DNA Blood and Tissue kit (Qiagen, Cat. No. 569504, Hilden, Germany) using the protocol for Gram-negative bacteria. The DNA was quantified using a Nanodrop 2000c spectrophotometer (ThermoFisher, Inc., Wilmington, DE, USA). Genomic DNA and PCR products were visualized on 1.5 and 4 % agarose gels, respectively, stained with ethidium bromide (0.33 mg/mL).
The detection of E. coli O157:H7 pathogenicity genes by mPCR
Standardization of the mPCR assay for pathogenicity genes was performed using specific primers for the stx1 (180 bp), stx2 (255 bp), eaeA (384 bp), and rfB (1000 bp) genes (Table 2). The reaction mixture was performed in a final volume of 25 µL, which consisted of 1X PCR buffer, 3 mM MgCl2, 1.25 U of Taq DNA polymerase (Invitrogen, Cat. No. 10342020, Brazil), 0.2 µM of each primer, 0.6 µM dNTPs (Invitrogen, Cat. No. 10297-018, Grand Island, NY, USA), 10-15 ng of DNA, and ultrapure water (Invitrogen, Cat. No. 10977-015) to adjust to the final volume. mPCR reaction conditions were as follows: an initial denaturation step at 94 oC for 5 min, followed by 35 cycles of denaturation at 94 oC for 30 s, annealing at 60 oC for 60 s, and extension at 72 oC for 75 s, with a final extension step of 72 oC for 7 min. Genomic DNA from E. coli ATCC 25922 was used as a negative control. Endpoint PCR reactions were performed in a thermocycler (Rosalind 24, Genes2Life SAPI de CV, Irapuato, Guanajuato, México).
Table 2 Primers used to detect pathogenicity and serogroup specific genes
| Primer | Sequence (5’- 3’) | Amplicon Size (bp) | Pathogenicity / Serogroup gene |
Reference |
|
stx1F stx1R |
ATAAATCGCCATTCGTTGACTAC AGAACGCCCACTGAGATCATC |
180 | Shiga Toxin 1 | (33) |
|
stx2F stx2R |
GGCACTGTCTGAAACTGCTCC TCGCCAGTTATCTGACATTCTG |
255 | Shiga Toxin 2 | |
|
eaeAF eaeAR |
GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG |
384 | Intimin (eaeA) |
|
|
rfBF
rfB R |
TAAGTAATGGAACGGTTGCTCT
CCCCACTCGTAAAATCCATC |
1000 | rfB | (34) |
|
fliCH4F fliCH4R |
ACGGCTGCTGATGGTACAG CGGCATCCAGTGCTTTTAAC |
244 | Flagellar antigen (fliCH4) |
(9) |
F=Forward; R=Reverse; wzx= (O-antigen flippase)
Amplicons of the stx2 gene were purified using a PCR purification kit (Qiagen, Cat. No. 28104, Hilden, Germany) and bidirectionally sequenced using an ABI PRISM 3100 instrument. The stx2 gene sequences were deposited in the GenBank; JA31 (MH040786), JA49 (MH040787), and JA50 (MH040788).
Identification by PCR of the fliCH4 gene
The methodology described by Paddock et al9 was performed to amplify the flagellar antigen, fliCH4 (244 bp), with the following modified PCR protocol: an initial denaturation step at 94 oC for 5 min, followed by 35 cycles of denaturation at 94 oC for 30 s, annealing at 57 oC for 45 s, extension at 72 oC for 30 s, and a final extension step at 72 oC for 7 minutes.
Phylogenetic identification using the stx2 gene sequence
Molecular identification was performed using the stx2 gene sequences obtained from the meat sample isolates. Sequence files were analyzed using SnapGene software (version 4.0.4; SnapGene GLS Biotech LLC, USA). The phylogenetic analysis was performed out using MEGA software version 6.035 with a partial region of the stx2 gene (accession number NZ_CP008957.1; 1 355 571-1 355 825 nt) to generate a phylogenetic tree, using reference sequences for stx2 obtained from GenBank (NCBI). The alignment was performed with the Clustal-W software of MEGA 6.0 and was edited using FigTree v1.4.1. The evolutionary distance was calculated using the Kimura 2 model. The phylogenetic relationship was estimated using the maximum likelihood method. The tree was inferred from 1000 replications (bootstraps).
Susceptibility to antibiotics
Antibiotic tests were performed on isolates positive for the stx2 and fliCH4 genes using two techniques. First, the Kirby-Bauer method with Mueller Hinton broth (Difco, Cat. No. 275730, Le Pont de Claix, France) and Mueller Hinton agar (MCD, Cat. No. 7131, Le Pont de Claix, France) was used in accordance with the disk diffusion assay method.36 Diffusion disks impregnated with tetracycline (30 µg), trimethoprim/sulfamethoxazole (25 µg, 1:19 w/w) 18, penicillin G (10 IU), oxacillin (1 µg), or clarithromycin (15 µg) (Oxoid, Cat. Nos. CT0043B, CT0159B, CT0054B, CT0693B, and CT0052B, respectively) were used. E. coli O157:H7 was used as a test control. The Kirby scale was used to evaluate susceptibility/resistance to antibiotics by measuring the growth inhibition halos of the bacterial isolates.36 The criteria used to read the inhibition halos were as follows: if the halo was ≥ 16-mm isolates were susceptible, if the halo was between 11-15-mm susceptibility was considered intermediate, and if the halo was ≤ 10 mm37 isolates were considered resistant to that specific antibiotic. The assay was performed in triplicate and the diameter of the growth inhibition area around the disks (mm) was determined after 18 h of incubation at 37 oC.
Additionally, a second methodology to confirm susceptibility/resistance to these antibiotics was used. An analysis of minimum inhibitory concentration (MIC, µg/mL) was carried out with 68 Microscan GN systems according to the Clinical and Laboratory Standard Institute (CLSI).
Results and discussion
Isolation of O157 presumptive isolates
This study focused on establishing the presence of STEC O157:H7. Table 1 shows that presumptive isolates from pork samples from the TIF slaughterhouse were low compared to isolates found in the non-TIF slaughterhouse, this is possibly due to higher temperatures used in the cold and cutting rooms of the non-TIF (0 and 25 oC) than in those of the TIF (-25 to -30; 4 to 8 oC) slaughterhouse.
There was a tendency to find a higher number of isolates in the pork and beef samples from the cut rooms, mainly in the non-TIF samples. The number of isolates was highest from pork loins in both the TIF and non-TIF slaughterhouses. Beef samples in the non-TIF slaughterhouse were also highest in the loin cuts compared to the pulp or rib cuts.
However, even when 224 presumptive isolates were obtained as presumptive STEC O157:H7 we were unable to confirm their identity by PCR as belonging to this serotype (described in the following section). After this finding, the study was re-focused on the characterization of these E. coli non-0157 isolates (Table 1). There are few reports on STEC in Mexico; however, studies of ground beef samples from Culiacan, Sinaloa27 and from supermarkets in the city of Puebla, Puebla26 did not find E. coli O157:H7, which was consistent with our findings.
Molecular characterization of O157 presumptive isolates
Standardization of the mPCR assay for pathogenicity genes is shown in Figure S1. Multiplex PCR showed that the presence of the stx2 pathogenicity gene was 1.40 % (3/224). Isolates that were positive for this gene were JA31, obtained from a beef loin sample from a non-TIF slaughterhouse and isolates JA49 and JA50, obtained from beef pulp samples from the same non-TIF slaughterhouse (Table 3, Figure S2). All three isolates were from beef and together showed 2.9 % (3/105) for the stx2 gene. Positive amplicons for stx2 were sequenced to confirm their identity. The sequences obtained for stx2 from isolates JA31, JA49, and JA50 were identical, so a single sequence was selected to build the phylogenetic tree (Figure 1), which shows that some O157:H7 strain cluster together with strains from serogroups O104:H4 and O145:H28 on a different branch of the tree, but that these strains are closer to the branch where JA31 is located than to the branch comprising other nonpathogenic Escherichia species. Phylogenetic analysis confirmed that the isolates from which the stx2 gene was positively amplified belonged to the stx2 gene from other E. coli serogroups reported.
Table 3 Escherichia coli genes detected in isolates from beef and pork
| Genes | ||||||
| Controls | rfB | stx1 | stx2 | eaeA | fliCH4 | wzx O157 |
| O157:H7 | + | + | + | + | - | + |
| ATCC 25922 | - | - | - | - | - | - |
| Isolates | ||||||
| JA31 | - | - | + | - | + | - |
| JA49 | - | - | + | - | - | - |
| JA50 | - | - | + | - | - | - |
| JA107 | - | - | - | - | + | - |
| JA113 | - | - | - | - | + | - |
| JA116& | - | - | - | - | + | - |
All isolates were gram-negative short bacilli; & isolated from pork.
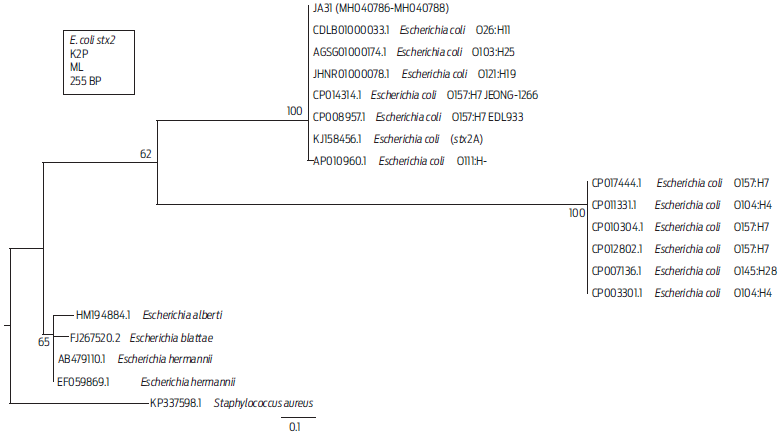
Figure 1 Distance tree derived from partial sequences of the stx2 gene. The tree was built with Mega 6 software, using the Kimura 2 parameter (K2P) substitution model. Branches show the percentages corresponding to 1 000 bootstrap replicates. GenBank accession numbers are found at the start of each name. The representative sequences of the three isolates from this assay are designated as JA31 (MH040786-MH040788). The scale bar indicates a genetic distance of 0.1 for each nucleotide substitution per site of the stx2 sequence. The KP337598.1 sequence from S. aureus was used as an outgroup to root the tree. ML= Maximum likelihood.
In the phylogenetic tree, the isolate JA31 was clustered with pathogenic strains that have caused clinical outbreaks (Figure 1). The KJ158456.1 sequence belongs to a strain from a patient with bloody diarrhea and was identified as nonmobile E. coli O59:H-, which possesses the stx2 gene, as well as other genes for type IV aggregated adherent fimbriae.37 Isolate JA31 was also clustered with the AGSG01000174.1 sequence from an entero-hemorrhagic E. coli (EHEC) strain of STEC O103:H25 that produced a severe outbreak that caused HUS due to contaminated food in Norway in 2006.15
Stx2 was the most frequently found gene in studies of E. coli non-O157 in ground beef samples collected from Chile.22 The stx2 toxin is 400 times more lethal than stx1 and plays a critical role in HUS development.38 There are reports of outbreaks associated with other serogroups that cause HUS39,40 in countries such as Argentina and Germany6,37,41 by pathogens such as E. coli O104:H4, which has the stx2 and fliCH4 genes, but does not possess the stx1, eaeA, or rfB genes.42 Based on this information, we decided to analyze our isolates for the presence of the fliCH4 gene characteristic of the O104:H4 serogroup9 (Table 3).
Figure S3 and Table 3 show that isolates JA31 (beef loin), JA107 (beef ribs), JA113 (beef ribs) and JA116 (pork pulp), all from the non-TIF slaughterhouse, were positive for the fliCH4. These data indicated 2.9 % (3/103) and 1.2 % (1/82) presence of the fliCH4 gene in beef and pork samples, respectively. The isolate JA31 in this assay, from the non-TIF slaughterhouse beef loin, had two genes, stx2 and fliCH4, to serogroup O104:H4, which shows characteristics of both enteroaggregative E. coli (EAEC) and enterohemorrhagic (EHEC).37 However, the wzx gene-specific for O104 was not amplified by PCR in this isolate, so we disregarded this possibility.
In this region of Mexico, there are no reports of E. coli containing the stx2 or fliCH4 genes from beef samples. Our group has recently reported the presence of E. coli O157:H7 in fecal samples of asymptomatic cattle in Sonora.43 Hence, conducting an in-depth study of JA31 to help understand the reason for the presence of these genes on a local scale is relevant. This knowledge may help understand E. coli disease outbreaks both locally and globally, possibly helping prevent global problems caused by STEC in the future.
In Mexico, the presence of E. coli O157:H7 has been reported in beef and pork carcasses in Northern Mexico, with an incidence of 66.6 % (12/18).24 In another work, the incidence of E. coli O157:H7 (2.7 %) and non-O157 (5 %) strains were reported from 258 samples taken from beef carcasses in a non-TIF slaughterhouse in Guadalajara. In this work, the incidence of E. coli non-O157 was only 1.40 % (out of 224 samples) from beef and pork carcasses. This rate was lower than those reported at the non-TIF slaughterhouse in Guadalajara.44
The JA31 isolate possessed the stx2 and fliCH4 genes. Within seropathotype D, there are O113:H4 and O7:H4 strains, and JA31 may be a candidate to demonstrate the presence of the wzx gene in serogroup O7 because we ruled out the possibility that JA31 is an O113 since this strain did not amplify with wzx genes for O113 (data not shown). The presence of these genes (stx2 and fliCH4) in the isolates in this study does not guarantee the expression and production of the Shiga toxin. Thus, further confirmation by evaluating the presence of the toxin in these isolates is needed.
Susceptibility and/or resistance to antibiotics
The isolates identified as E. coli that tested positive for the stx2 gene (3/224) and/ or the fliCH4 gene (4/224), displayed resistance to tetracycline, oxacillin, penicillin G, ampicillin, sulbactam, cefazolin and trimethoprim/sulfamethoxazole while being susceptible to the other 16 antibiotics tested (Table 4). The main resistance mechanism displayed by these isolates belongs to beta-lactamase inhibitors, and tetracyclines affecting the bacterial protein synthesis.45 Such multiple-resistance to antibiotics has been previously reported in STECs.46
Table 4 Antibiotic susceptibility of Escherichia coli isolates with stx2 or fliCH4 genes from meat products
| Method | Antibiotic | Dose per disk | JA31 | JA49 | JA50 | JA107 | JA113 | JA116 |
| Minimum inhibitory concentration **MIC (µg/mL) |
AK | s < 16 | s < 16 | s < 16 | s < 16 | s < 16 | s < 16 | |
| Aug | s < 8/4 | s < 8/4 | s < 8/4 | s < 8/4 | s < 8/4 | s < 8/4 | ||
| A/S | r > 16/8 | r > 16/8 | r > 16/8 | r > 16/8 | r > 16/8 | r > 16/8 | ||
| AM | r > 16 | r > 16 | r > 16 | r > 16 | r > 16 | r > 16 | ||
| Cfz | r > 4 | r > 4 | r > 4 | r > 4 | r > 4 | r > 4 | ||
| Cpe | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | ||
| Cft/CA | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | ||
| Cft | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | ||
| Caz | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | ||
| Caz/CA | s < 0.25 | s < 0.25 | s < 0.25 | s < 0.25 | s < 0.25 | s < 0.25 | ||
| Cax | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | ||
| Crm | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | ||
| Cp | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | ||
| Etp | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | s < 0.5 | ||
| Gm | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | ||
| Imp | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | ||
| Lvx | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | ||
| Mer | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | s < 1 | ||
| P/T | s < 16 | s < 16 | s < 16 | s < 16 | s < 16 | s < 16 | ||
| Te | r > 8 | r > 8 | r > 8 | r > 8 | r > 8 | r > 8 | ||
| Tgc | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | s < 2 | ||
| To | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | s < 4 | ||
| T/S | r > 2/38 | r > 2/38 | r > 2/38 | r > 2/38 | r > 2/38 | r > 2/38 | ||
| Kirby-Bauer (Inhibition halos in mm) |
Te | 30 µg | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) |
| FOS | 200 µg | ≥16 (s) | ≥16 (s) | ≥16 (s) | ≥16 (s) | ≥16 (s) | ≥16 (s) | |
| CLR*+ | 15 µg | 10.0 (r) | 12.9 (is) | 14.0 (is) | 0.0 (r) | 0.0 (r) | 0.0 (r) | |
| OX*+ | 1 µg | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | |
| P*+ | 10 iu | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | 0.0 (r) | |
** 68 Microscan GN systems; MIC = minimum inhibitory concentration according to Clinical and Laboratory Standard Institute criteria. AK, amikacin; Aug, amoxicillin-clavulanate; A/S, ampicillin-sulbactam; AM, ampicillin; Cfz, cefazolin; Cpe, cefepime; Cft/CA, cefotaxime-clavunalate; Cft, cefotaxime; Caz, ceftazidime; Caz/CA, ceftazidime- clavunalate; Cax, ceftriaxone; Crm, ceforoxime; Cp, ciprofloxacin; ETP, ertapenem; Gm, gentamicin; Imp, imipenem; Lvx, levofloxacin; Mer, meropenem; P/T, piperacillintazobactam; Te, tetracycline; TGC, tigecycline; To, tobramycin; T/S, trimethoprim-sulfamethoxazole; Te, tetracycline; T/S, trimethoprim/sulfamethoxazole; FOS, fosfomycin; CLR, clarithromycin; OX, oxacillin; P, penicillin G; IU= international unit.
* Isolates are susceptible (S) to an antibiotic when they show an inhibition halo ≥ 16 mm, intermediate susceptible (IS) when they show an inhibition halo between 11 and 15 mm, and resistant (R) to an antibiotic if the halo is ≤ 10 mm. +antibiotic not included in the Clinical and Laboratory Standards Institute for Enterobacteriaceae.
The combination of antibiotics is recommended to treat a range of human infections, including urinary tract infections, uncomplicated acute cystitis, and pyelonephritis. Nevertheless, the use of antibiotics in individuals with STEC infections is controversial and generally not recommended, this is because some antibiotics affect DNA replication by activating the SOS response in bacteria and indirectly increasing the number of toxins such as ST.17,18 This study confirms the variability concerning the susceptibility or resistance to antibiotics of E. coli strains containing the stx2 and/or fliCH4 genes.
Conclusions
This study reports that no E. coli STEC O157:H7 was found in 224 fresh beef and pork meat samples taken from one TIF and one non-TIF slaughterhouse and 12 local butcheries in Ciudad Obregon, Sonora. Nevertheless, the stx2 and fliCH4 genes were present in some isolates from beef meat at low frequencies (1.40 and 1.78 %). Isolates containing the stx2 and/or fliCH4 gene showed multiple antibiotic resistance to tetracycline, penicillin G, oxacillin, ampicillin, sulbactam, cefazolin and trimethoprim/sulfamethoxazole.
Data availability
The partial region of the stx2 gene were deposited in the NCBI database with accession number NZ_CP008957.1; 1 355 571-1 355 825 nt.
Acknowledgments
JAAJ received Ph.D. fellowships from the Consejo Nacional de Ciencia y Tecnología (CONACyT) (Mexico). The E. coli O157:H7 strain was kindly donated by Dr. Gracia Gómez-Anduro from CIBNOR, Mexico. We thank Dr. Brandon Loveall of improvence for English proofreading of the manuscript.
Conflicts of interest
The authors have no conflict of interest to declare in regard to this publication.
Author contributions
Conceptualization: EU Cantu-Soto, IE Maldonado-Mendoza, AM Figueroa-Lopez.
Formal analysis: JA Anduro-Jordan, EU Cantu-Soto, IE Maldonado-Mendoza, AM Figueroa-Lopez.
Funding acquisition: EU Cantu-Soto, IE Maldonado-Mendoza, AM Figueroa-Lopez.
Investigation: JA Anduro-Jordan.
Methodology: JA Anduro-Jordan.
Project administration: EU Cantu-Soto, IE Maldonado-Mendoza, AM Figueroa-Lopez.
Supervision: EU Cantu-Soto, IE Maldonado-Mendoza, AM Figueroa-Lopez.
Validation: EU Cantu-Soto.
Writing-original draft: JA Anduro-Jordan.
Writing-review and editing: EU Cantu-Soto, AM Figueroa-Lopez, IE Maldonado-Mendoza, JA Anduro-Jordan.











 text new page (beta)
text new page (beta)



