Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Abanico veterinario
On-line version ISSN 2448-6132Print version ISSN 2007-428X
Abanico vet vol.13 Tepic Jan./Dec. 2023 Epub Oct 27, 2023
https://doi.org/10.21929/abavet2023.14
Literature review
Review: SARS-CoV-2 natural infection in animals
1Departamento de Ciencias Veterinarias, Universidad Autónoma de Aguascalientes. México.
2Departamento de Microbiología, Universidad Autónoma de Aguascalientes. México.
The objective of this study was to evaluate epidemiological and pathological reports of confirmed outbreaks and cases of zoo, farm, and pet animals naturally infected with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Confirmed reports of outbreaks and cases of SARS-CoV-2 infected animals were obtained by systematically researching and analyzing information available in open access databases. The reports were evaluated, incorporated into a database, classified, and integrated to estimate average morbidity and mortality values and characteristic clinical manifestations of SARS-CoV-2 in animals. Postmortem lesions and microscopic alterations are also included. Diagnostic and therapeutic alternatives to confirm or mitigate infection in animals are outlined. Finally, control methods include isolation, culling of affected populations, and the development of the first available vaccines for animals. Available information suggests that domestic and zoo animals have become accidental hosts of SARS-CoV-2, becoming infected primarily through contact with COVID-19 affected humans; although there is concern that animals could become a risk for maintaining and spreading new mutant variants of the virus, which could eventually return to human populations.
Keywords: SARS-CoV-2; COVID-19; zoonosis; animal infection
El objetivo de este estudio fue evaluar los informes epidemiológicos y patológicos de brotes y casos confirmados de la enfermedad por SARS-CoV-2 en animales de zoológico, granja y mascota infectados naturalmente. Se obtuvieron informes de brotes y casos de animales infectados por SARS-CoV-2 mediante búsqueda, análisis y clasificación sistemática de información disponible en bases de datos de libre acceso. Los informes se evaluaron e incorporaron a una base de datos, se clasificaron e integraron para identificar los valores característicos de morbilidad, mortalidad y manifestaciones clínicas del SARS-CoV-2 en animales; además de lesiones post mortem y alteraciones microscópicas. Se indican alternativas diagnósticas y terapéuticas para confirmar o mitigar la infección en animales; métodos de control como el aislamiento, sacrificio de poblaciones afectadas y las primeras vacunas disponibles para animales. La información disponible sugiere que los animales se han convertido en huéspedes accidentales del SARSCoV-2, infectándose principalmente a través del contacto con personas afectadas por COVID-19; aunque existe la preocupación de que los animales podrían convertirse en un riesgo de mantenimiento y propagación de nuevas variantes virales, que eventualmente podrían volver a las poblaciones humanas
Palabras clave: SARS-CoV-2; COVID-19; zoonosis; infección animal
Introduction
In December 2019, in Wuhan, Hubei province, China, the first cases of sick patients with pneumonia of unknown origin were epidemiologically linked to the Huanan Seafood Market (Ciotti et al., 2020; Xie & Guangzhou, 2020), where more than one hundred different species of animals were sold, including mammals (civets, koalas, bats, foxes), birds (ostriches, pheasants, ducks, peacocks), reptiles, and amphibians (crocodiles, snakes, frogs) in overcrowded conditions, poor hygiene, and inadequate handling of water, waste, and cadavers (Fasina, 2020; Jalava, 2020; Ji et al., 2020b).
It was quickly identified that this disease was caused by a new ß-coronavirus initially named new coronavirus disease (2019-nCoV). On January 12, 2020, it was officially named coronavirus disease 2019 (COVID-19) by the WHO (World Health Organization) or SARS-CoV-2 by the International Committee on Viral Taxonomy (Ahn et al., 2020). The first reports in humans indicate that they were in contact with civets (Paguma larvata) sold in the Huanan Seafood Market before manifesting the first clinical signs of the disease, establishing a possible zoonotic origin of the disease. However, several studies indicate that this civet species is only an intermediate host because the genetic sequence of SARS-CoV-2 showed a 96.2% homology with the bat coronavirus CoV-RaTG13, bats being the species responsible for the outbreak of this new disease Salata et al., 2019; Ye et al., 2020).
This is not the first time that a species of β-coronavirus of animal origin has affected humans; in 2003, in the province of Guangdong, China, a coronavirus originating in bats affected humans through an intermediate host (palm civet; Paguma larvata). This virus is called severe acute respiratory syndrome (SARS) and caused 916 deaths and affected 8,422 people with a mortality of 10.9% (Singhal, 2020). In 2012, in Saudi Arabia, another virus originated in bats called Middle East respiratory syndrome coronavirus (MERS-CoV) affecting 2,494 people. It caused 858 deaths with a mortality of 34% (Shereen et al., 2020). Reports of SARSCoV-2 outbreaks in animals are scarce, but their geographical distribution is very diverse. This coincides with a worldwide spread of the virus. Therefore, this paper aims to review the presence of the infection in pets, farm animals and zoos. Information on the origin and parallel spread between humans and animals, the mechanisms of infection and its impact on animal health are reviewed. Viable alternatives for diagnosis and therapy of SARSCoV-2 in animals are presented, and control of the pandemic with available vaccines is highlighted. The objective of this study was to evaluate reports of outbreaks and confirmed cases of animals naturally infected with SARS-CoV-2 about epidemiological and pathogenic patterns in zoo, farm, and domestic animals.
Methods
This review was conducted by establishing a research objective, search strategies, and relevant research articles and reports; selection of bibliographic material, data extraction, data mapping, and summary of results were performed. The literature for this review was identified by searching online databases (World Organization for Animal Health, OIE, and World Health Organization, WHO, Google scholar, PubMed, and Web of Science). We searched for scientific publications from 2019 to 2022. The search terms were 'CORONAVIRUS', 'SARS', 'ANIMAL' and "COVID-19". All relevant scientific publications and official reports were included in the review, but other types of information (congresses, theses, etc.) were excluded from the analysis. Two investigators independently evaluated each bibliographic source. The two sets of selected literature were then compared; disagreements about the inclusion of the literature were resolved by group discussion to make the decision. Data on design, objectives, animal population, instrumental methodology, main results, and conclusions were extracted. Articles were categorized into the following areas "Origin", "Host species", "SARS-CoV-2", "Pathogenesis", "Clinical findings", "Lesions", "Therapeutic strategies", "Control" and "Vaccines". Quantitative data reported in the outbreaks were summarized and averages by animal species were estimated. All conclusions and statements in this review are based on published information, as indicated in the references.
Distribution of the new coronavirus
On December 30, 2019 (Chowdhury & Oommen, 2020; Rothan & Byrareddy, 2020) an outbreak of pneumonia of unknown etiology was reported in the city of Wuhan (Fig. 1). On January 7, 2020, the isolation and identification of the genome of a new coronavirus was performed (Bulut & Kato, 2020). On January 13, 2020, an increase in the propagation of the virus associated with nosocomial infections and direct contact with infected relatives was detected. On the same day, the first case of the novel coronavirus was confirmed in Thailand, and on January 19, the first cases were reported in Beijing, which indicated the spread of the virus within China and across the surrounding region (Sun et al., 2020). By January 22, the National Health Commission of China reported 17 deaths and 571 infected people in 25 provinces of China (Rothan & Byrareddy, 2020), which is why the government of Wuhan implemented a total closure of activities inside and outside the city. Unfortunately, these measures coincided with the beginning of the Chinese New Year, which meant that more than five million people left the city to return to their homes. This caused an increase in the number of confirmed cases; therefore, on January 30, 2020, the WHO declared the disease caused by SARS-CoV-2 a “Public Health Emergency of International Concern” (WHO, 2020).

Source: Own elaboration with data from: (OIE-WAHIS, 2021; WHO, 2021).
Figure 1 Timeline of the epidemiological spread of SARS-CoV-2 in humans and animals.
During the first week of February in Wuhan, a turning point in the epidemic was observed, where the daily number of confirmed cases began to decrease; however, it is not known whether the reduction in virus transmission was due to the established blockade, the public health measures implemented or other factors (Sun et al., 2020). Since its onset in China, the disease spread rapidly. The number of cases increased exponentially worldwide, and in a couple of months, it extended to all continents except Antarctica (Bulut & Kato, 2020; Chowdhury & Oommen, 2020; Rothan & Byrareddy, 2020; Sun et al., 2020; WHO, 2021).
Although there is extensive documentation on the progress of COVID-19 in humans, there are few studies related to the epidemiological distribution of SARS-CoV-2 in animals. In outbreaks in animals, the presence of clinical respiratory signs has been reported, in addition to low mortality and morbidity. It should be noted that contact with previously infected owners or workers has been identified as a source of infection for animals (OIEWAHIS, 2021).
Epidemiological information worldwide indicates a greater number of outbreaks in domestic animals, with morbidity and mortality rates of 52.5% and 2.6%, respectively. Cats are the species with the most reported outbreaks (OIE-WAHIS, 2021). In zoo animals, the disease has been identified in gorillas, cougars, lions, leopards, and tigers, with a presence of respiratory signs in 83.7% of the outbreaks (OIE-WAHIS, 2021).
One year after the beginning of the pandemic, on December 8, 2020, the first person worldwide was vaccinated against SARS-CoV-2, and a new viral variant was identified in the United Kingdom. By January 2021, two million deaths and more than 100 million infected people were counted worldwide; the OIE established a risk manual for farm animals due to the great losses in the production of minks (OIE-WAHIS, 2021). By April, the number of deaths in humans increased to three million, and the application of the first vaccines against COVID-19 in animals began. In August and October, the first reports of COVID-19 in wild deer, otters, and binturong were presented, reporting a total of 1,850 confirmed cases in animals and 267 million in humans. A possible animal-to-human transmission of SARS-CoV-2 was documented on January 31, 2022, in Hong Kong, where the Delta variant was detected in hamsters as well as in a pet store employee (OIEWAHIS, 2021) .
Possible hosts of the virus
In China, there are established farms dedicated to the breeding of exotic animals for human consumption (civets, bats, pangolins, snakes), which distribute their products to various restaurants. Consequently, it has been assumed that farms, restaurants, and wet markets were probably responsible for the zoonotic origin of the new coronavirus (Tiwari et al., 2020). Different authors point to four possible animal species as the initial hostsbats, pangolins, civets, and snakes (Fig. 2) due to the high similarity in their genome and in some surface proteins that the coronaviruses of these animal species have in relation to SARS-CoV-2 (Anand et al., 2020; Ji & Li, 2020; Lau et al., 2020; Zheng, 2020).
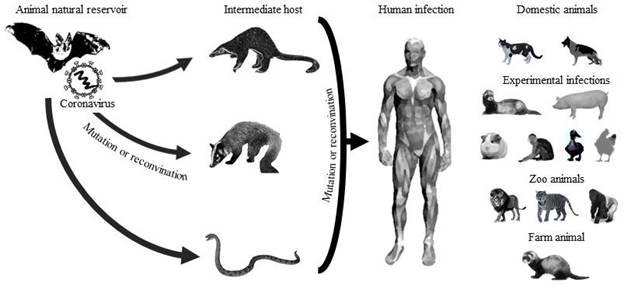
Source: Own elaboration with data from: (Anand et al., 2020;Ji & Li, 2020;Lau et al., 2020;Zheng, 2020).
Figure 2 There is a possible origin, transmission, and infection of SARS-CoV-2. The natural reservoir is bats, and the possible intermediate hosts are pangolins, civets, and snakes. Species infected with SARSCoV-2 include humans, domestic animals (dogs and cats); animals with experimental infections (ferrets, pigs, macaques, ducks, and chickens), zoo animals (lions, tigers, and gorilla), and farm animals (minks).
It has been described those bats are reservoirs for different species of coronavirus due to because when the carry the virus, it remains latent and asymptomatic. As well, the great distances they must travel to obtain food allows them to infect other animal species (Tiwari et al., 2020). Laboratory findings reported that SARS-CoV-2 has a genetic identity of 87.6%, 87.8%, and 96.1% with SARSr-Rp-BatCoV-ZXC21, SARSr-Rp-BatCoV-ZC45, and SARSr-Ra-BatCoV-RaTG13, respectively, which are viruses identified in bats (Rhinolophus pusillus, R affinis) captured in China (Lau et al., 2020).
Although SARS-CoV-2 appears to have originated initially in bats, it is probably that it used pangolins as intermediate hosts. Analysis of pangolin samples revealed several viral sub-lineages related to SARS-CoV-2 (Table 1) (Hu et al., 2021; Zheng, 2020). Several studies point to the Malayan pangolin (Manis javanica) as a possible natural reservoir or intermediate host of COVID-19 (Zhang et al., 2020). Genomic analyses suggest that GD Pangolin CoV has a nucleotide sequence similarity of 90.32% and 90.24% with SARSCoV-2 and Bat-CoV-RaTG13, respectively. However, the similarity in some viral protein sequences could be 100% (Liu et al., 2020). These results could consider pangolins as an intermediate host of SARS-CoV-2 (Xiao et al., 2020), referring that it probably could have originated from a genetic recombination between bat and pangolin coronaviruses (Wong et al., 2020).
Table 1 SARS-CoV-2-related coronaviruses and their lineages in Sarbecovirus-infected animals
| Bat | Pangolin | Human | |
|---|---|---|---|
| SARS-CoV | Bat SARSr-CoV Rs4231 | SARS-CoV GZ02 | |
| Bat SARSr-CoV SHC014 | SARS-CoV Tor2 | ||
| Bat SARSr-CoV WIV1 Bat SARSr-CoV Rp3 Bat SARSr-CoV HKU3-1 | SARS-CoV BJ01 | ||
| SARS-CoV-2 | Bat coronavirus ZXC21 | GD_Pangolin-CoV | Alpha variant (United Kingdom) |
| Bat coronavirus ZC45 | GX_Pangolin-CoV_P2V | SARS-CoV-2 B.1.1.7 | |
| Bat coronavirus RmYN02 | GX_Pangolin-CoV_P5E | Beta variant (South Africa) | |
| Bat coronavirus RaTG13 | GX_Pangolin-CoV_P1E | SARS-CoV-2 B.1.351 | |
| GX_Pangolin-CoV_P5L | SARS-CoV-2 B.1.351.2 | ||
| GX_Pangolin-CoV_P4L | SARS-CoV-2 B.1.351.3 | ||
| GX_Pangolin-CoV_P3B | Gamma variant (Brazil) SARS-CoV-2 P.1 SARS-CoV-2 P.1.1 SARS-CoV-2 P.1.2 Delta variant (India) SARS-CoV-2 B.1.617.2 SARS-CoV-2 AY.1 SARS-CoV-2 AY.2 Omicron variant (South Africa) SARS-CoV-2 B.1.1.529 |
Source: Own elaboration with data from: (Hu et al., 2021).
However, other researchers consider civets as a possible intermediate host and transmitter of the new coronavirus due to seroepidemiological results that identified antibodies against SARS-CoV-2 in people previously exposed to contact with these animals (Salata et al., 2019). Phylogenetic analysis of SARS-CoV-2 showed a genome similarity of 78.6% with Civet-SARSr-CoVs. The ability of SARS-CoV-2 to emerge and infect humans could be caused by the genetic similarity between SARSr-BatCoV and Civet-SARSr-CoV from Yunnan, China (Lau et al., 2020).
Similarly, there is a theory that the snake can serve as a natural reservoir of SARS-CoV2 and participate in its transmission between animals and humans (Ji & Li, 2020), due to the similarity of the codons of SARS-CoV-2, bat-SL-CoVZC45, and snake coronavirus. However, SARS-CoV-2 has not been isolated in snakes so far (Zheng, 2020). These investigations suggest that COVID-19 possesses similar genetic information to bat and snake coronaviruses, and thus, recombination in its viral binding proteins could be responsible for cross-species transmission (Ji et al., 2020a).
Infected animal species
The identification of reservoir animals as infectious agents with zoonotic potential is essential for the establishment of sanitary measures to mitigate the adverse effects of the disease. However, this type of infectious agent can infect a wide variety of domestic or wild animals (OIE-WAHIS, 2021). Because coronaviruses have a wide diversity of animal species they can infect, there is the possibility they can cross the barrier between species by means of genetic recombination mechanisms that allow them to infect other animal species and even humans (Abdel-Moneim & Abdelwhab, 2020).
This process has been previously identified in SARS and MERS outbreaks. Like SARSCoV-2, these were caused by a coronavirus that jumped the species barrier. Although the true origin of SARS-CoV-2 is currently unknown, the phylogenetic identity has served as a basis for suspicions that it was initially transmitted by bats, where this virus, in addition to infecting humans, has infected other animal species (Tiwari et al., 2020). Other research detected probable SARS-CoV-2 deer-to-deer transmission that demonstrates the possibility of opening new evolutionary pathways (Hale et al., 2022).
Pet animals
The first reported case was identified in Hong Kong on February 29, 2020, in a domestic dog belonging to a family infected with coronavirus, whereas in domestic and zoo felids, the first outbreaks of this disease were reported on March 28 and April 6, 2020 (OIEWAHIS, 2021), respectively. The first reports of COVID-19 in dogs and cats in China were diagnosed by using polymerase chain reaction (PCR) for the isolation and sequencing of the viral genome (Ruiz-Arrondo et al., 2020; Sit et al., 2020). Studies showed that after a local outbreak in humans, antibodies to SARS-CoV-2 were detected in dogs and cats (Abdel-Moneim & Abdelwhab, 2020). After the outbreak in China, cases of COVID-19 were reported in domestic animals in different countries of the world (OIE-WAHIS, 2021). In all the outbreaks, the animals belonged to people previously ill with COVID-19. This evidence suggests a possible origin of the infection: the transmission from human to animal as well as the transmission from infected animals to humans, which has not been identified until now (Sit et al., 2020).
Farm animals
The first official report on SARS-CoV-2 in farm animals comes from mink farms located in the Netherlands. On April 19, 2020, some animals began to manifest clinical respiratory signs ranging from a slight nasal discharge to severe respiratory distress. Subsequently, outbreaks have occurred in other mink farms, affecting a population of 736,257 animals. The possible cause of infection was contact with sick workers with COVID-19. Due to the growing number of infected animals and the discovery of a new mutation of SARS-CoV2, several countries of the European Union proceeded to slaughter more than 17 million minks, thus preventing the propagation and dispersion of a new viral variant (OIE-WAHIS, 2021; Oreshkova et al., 2020).
Zoo animals
There have been reports of lions, tigers, cougars, leopards, lynx, otters, coati, binturong and gorillas diagnosed by PCR and genetic sequencing as SARS-CoV-2-positive. Most of these animals presented respiratory clinical signs where the possible cause of infection was previously infected zookeepers (OIE-WAHIS, 2021).
Experimental infections
Experimental infections in various animal species have been developed in the laboratory to understand the pathogenesis and development of the disease. For this reason, species such as hamsters, ferrets, macaques, shrews, and mice have been inoculated by different routes (intratracheal, intranasal, ocular, and oral), where the immunopathology, transmission and development of COVID-19 treatments have been established (Abdel-Moneim & Abdelwhab, 2020; Shi et al., 2020). Experimentally infected swine and poultry (quails, geese, ducks, turkeys, and chickens) showed no clinical signs or pathological lesions, and it was not possible to identify the presence of viral RNA or antibodies. These results indicate that none of these animal species are susceptible to SARS-CoV-2, and they do not play an important role in its transmission (Shi et al., 2020; Suarez et al., 2020).
Characteristics of SARS-CoV-2
Until recently, coronaviruses in humans were not considered to cause anything more serious than the common cold. However, before advent of the COVID-19 pandemic, these viruses have had a higher profile in veterinary medicine (Table 2). SARS-CoV-2 is a βcoronavirus of the subgenus Sarbecovirus, subfamily Orthocoronavirinae, whose members primarily infect bats (Wrobel et al., 2020). The Coronaviridae family consists of four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Chen et al., 2021).
Table 2. Taxonomic classification of the coronaviruses
| Subfamily | Genus | Subgenus | Species |
|---|---|---|---|
| Letovirinae | Alphaletovirus | Milecovirus | Microhyla letovirus 1 |
| Orthocoronavirinae | Alphacoronavirus | Duvinacovirus | Human coronavirus 229E |
| Minacovirus | Mink coronavirus 1 | ||
| Pedacovirus | Porcine epidemic diarrhea virus | ||
| Setracovirus | Human coronavirus NL63 | ||
| Tegacovirus | Alphacoronavirus 1 (Canine, feline and porcine coronavirus, Transmissible gastroenteritis virus) | ||
| Betacoronavirus | Embecovirus | Human coronavirus HKU1 | |
| Merbecovirus | Middle East respiratory syndrome-related coronavirus | ||
| Sarbecovirus | Severe acute respiratory syndromerelated coronavirus | ||
| Deltacoronavirus | Buldecovirus | Coronavirus HKU15 (Porcine) | |
| Gammacoronavirus | Igacovirus | Avian coronavirus (infectious bronchitis) |
Source: Own elaboration with data from: (ICTV, 2021).
SARS-CoV-2 is an enveloped, capsid-bound, non-segmented, single-stranded RNA virus with a series of spikes on its surface, which are known as “S” proteins and are responsible for its binding to cellular epithelia (Majumder & Minko, 2021).
As a member of the Betacoronavirus genus, SARS-CoV-2 shares a genomic similarity of 50% and 79% with MERS-CoV2 and SARS-CoV, respectively, and SARS-CoV-2 encoded proteins have a similar length to those corresponding to SARS-CoV (Hu et al., 2021). Several authors point out the degree of molecular divergence between SARS-CoV-2 and other coronaviruses. The phylogenetic analysis of SARS-CoV-2 shows a relationship with SARS-CoV, and other SARS-related coronaviruses found in bats (Tang et al., 2020).
SARS- CoV-2 clusters in a distinct lineage along with four bat coronaviruses, RaTG13, RmYN02, ZC45, and ZXC21, as well as new coronaviruses recently identified in pangolins, which cluster similarly to SARS coronavirus (Hu et al., 2021).
Pathogenesis
The routes of transmission identified in animals for infection caused by COVID-19 have been reported as the main cause of contact with infected persons as well as the inhalation of aerosol particles (OIE-WAHIS, 2021), which have the capacity to lodge in the respiratory tract (Rothan & Byrareddy, 2020; Woodby et al., 2020). In animals, a incubation period has been described in experimental and natural SARS-CoV-2 infections (2 to 14 days; Fig. 3) (Munster et al., 2020; Shi et al., 2020; Tiwari et al., 2020). It has been observed in most outbreaks that the onset of clinical manifestations and recovery is earlier, with the first clinical signs being observed as early as 0-10 days postexposure (dpe) and ceasing between 9 or 17 dpe. However, the presence of viral RNA can be detected from 3-13 dpe, whereas the formation of antibodies is generated at 10 dpe or more. Furthermore, the presence of radiographic changes in the lungs and pathological lesions in various organs (1-3 dpe) has been described, which may persist after the end of the disease (Abdel-Moneim & Abdelwhab, 2020; Chan et al., 2020; Lu et al., 2020). Outbreaks with clinical manifestations have occurred mainly in animals with a history of cardiovascular, respiratory, neurological diseases, neoplasms, and obesity (OIE-WAHIS, 2021; Ruiz-Arrondo et al., 2020).

Source: Own elaboration with data from: (Abdel-Moneim & Abdelwhab, 2020; Chan et al., 2020; Lu et al., 2020)
Figure 3 Natural history of SARS-CoV-2 in animals.
Nasal epithelial cells have been identified as the site of initiation of SARS-CoV-2 infection, where an increase in ACE2 levels has been observed due to a high binding affinity for the SARS-CoV-2 spike glycoprotein. Subsequently, through inspiration, the virus manages to colonize the respiratory tract, where it invades type II pneumocytes and initiates a phase of rapid viral replication, producing localized inflammation, increased vascular permeability, increased chemotaxis, and pulmonary edema (Chan et al., 2020; Munster et al., 2020).
The presence of edema in the lungs hinders them from filling with air, causing low oxygen saturation, pneumonia, lung damage, and death. Lung damage allows the virus to enter the bloodstream and initiate the activation of the coagulation cascade, thrombus formation, and damage to other organs such as the heart, kidney, brain, stomach, and intestine (Lotfi & Rezaei, 2020; Shi et al., 2020).
In animals, reports of natural and laboratory infections have described the presence of histopathological lesions in the liver, heart, kidney, and intestine; respiratory failure; arrhythmias; and complication with opportunistic bacterial infections (Lu et al., 2020; OIEWAHIS, 2021). These reports demonstrate that if the organism is unable to control the early stages of viral replication, it can give rise to severe disease and cause death as a result of an impaired or delayed immune response (Woodby et al., 2020).
Clinical manifestations in animals
During the development of this pandemic, the clinical symptomatology in humans has been described in detail; however, in animals, there are currently few reports of outbreaks describing the clinical manifestations, lesions, and pathological alterations associated with SARS-CoV-2 infection (OIE-WAHIS, 2021). It has been observed that the presentation of the disease in animals (Table 3, 4) is very similar to its presentation in humans. In animals, respiratory clinical signs include cough, dyspnea, nasal secretion, sneezing, fever, and inflammation in the anterior and posterior respiratory tract (Abdel-Moneim & Abdelwhab, 2020; OIE-WAHIS, 2021; Shi et al., 2020).
Table 3 Analysis of SARS-COV-2 outbreaks in domestic, zoo, wild, and farm animals
| Animal species | Report date | Outbreaks (No.) | Outbreaks with clinical respiratory signs (%) | Susceptible animals (No.) | Apparent morbidity (%) | Deaths (No.) | Apparent mortality (%) | Confirmed animals (No.) | Probable source of contagion | |
|---|---|---|---|---|---|---|---|---|---|---|
| First | Last | |||||||||
| Domestic | 02/29/20 | 12/06/21 | 85 | 43.5 | 270 | 52.5 | 7 | 2.6 | 142 | Owner |
| -Cat | 03/28/20 | 12/06/21 | 43 | 46.5 | 140 | 55.0 | 3 | 2.1 | 77 | Owner |
| -Dog | 02/29/20 | 12/06/21 | 40 | 37.5 | 127 | 49.6 | 4 | 3.1 | 63 | Owner |
| -Ferret | 12/23/20 | 09/28/21 | 2 | 100 | 3 | 66.6 | 0 | 0.0 | 2 | Owner |
| Zoo | 08/11/20 | 10/28/21 | 49 | 83.7 | 223 | 47.0 | 2 | 0.8 | 105 | Staff |
| - Binturong | 10/14/21 | 10/14/21 | 1 | 100 | 1 | 100 | 0 | 0.0 | 1 | Staff |
| -Cougar | 08/11/20 | 03/18/21 | 4 | 25.0 | 16 | 18.7 | 0 | 0.0 | 3 | Staff |
| -Fisher cat | 10/14/21 | 10/14/21 | 1 | 100 | 1 | 100 | 0 | 0.0 | 1 | Staff |
| -Gorilla | 01/11/21 | 10/14/21 | 3 | 33.3 | 34 | 38.2 | 0 | 0.0 | 13 | Staff |
| -Lion | 01/11/21 | 12/02/21 | 15 | 92.3 | 68 | 54.4 | 1 | 1.4 | 37 | Staff |
| -Otter | 04/28/21 | 09/13/21 | 3 | 100 | 14 | 50 | 0 | 0.0 | 7 | Staff |
| -Coati | 10/14/21 | 10/14/21 | 1 | 100 | 1 | 100 | 0 | 0.0 | 1 | Staff |
| -Snow leopard | 12/18/20 | 10/28/21 | 5 | 100 | 13 | 76.9 | 1 | 7.6 | 10 | Staff |
| -Tiger | 12/23/20 | 12/07/21 | 15 | 93.3 | 75 | 41.3 | 0 | 0.0 | 31 | Staff |
| -Lynx | 01/12/21 | 01/12/21 | 1 | 100 | 1 | 100 | 0 | 0.0 | 1 | Staff |
| Wild deer | 08/31/21 | 12/01/21 | 2 | 0.0 | 360 | 3.0 | 0 | 0.0 | 11 | Wild |
| Farm mink | 04/26/20 | 12/07/21 | 33 | 24.2 | 736257 | 0.2 | 8601 | 1.1 | 1592 | Staff |
| America | 04/6/20 | 10/28/21 | 93 | 72.0 | 56404 | 0.6 | 8062 | 14.2 | 391 | |
| -Domestic | 04/22/20 | 09/21/21 | 50 | 60.0 | 210 | 45.7 | 7 | 3.3 | 96 | Owner |
| -Farm | 08/20/20 | 12/9/20 | 3 | 100 | 55657 | 0.36 | 8053 | 14.4 | 200 | Staff |
| -Zoo | 04/6/20 | 10/28/21 | 38 | 89.5 | 177 | 47.4 | 2 | 1.1 | 84 | Staff |
| -Wild | 08/31/21 | 12/01/21 | 2 | 0.0 | 360 | 3.0 | 0 | 0.0 | 11 | Wild |
| Europe | 03/28/20 | 10/25/21 | 58 | 32.8 | 680656 | 0.2 | 548 | 0.1 | 1427 | |
| -Domestic | 03/28/20 | 12/06/21 | 21 | 33.3 | 37 | 67.5 | 0 | 0.0 | 25 | Owner |
| -Farm | 04/26/20 | 12/07/21 | 30 | 16.7 | 680600 | 0.2 | 548 | 0.1 | 1392 | Staff |
| -Zoo | 12/8/20 | 12/07/21 | 7 | 100 | 19 | 52.6 | 0 | 0.0 | 10 | Staff |
| Asia | 02/29/20 | 12/02/21 | 16 | 18.8 | 45 | 62.2 | 0 | 0.0 | 28 | |
| -Domestic | 02/29/20 | 10/06/21 | 14 | 7.1 | 23 | 91.3 | 0 | 0.0 | 21 | Owner |
| -Zoo | 09/08/21 | 09/08/21 | 2 | 100 | 22 | 31.8 | 0 | 0.0 | 7 | Staff |
| Africa -Zoo | 08/11/20 | 07/27/21 | 2 | 100 | 5 | 80.0 | 0 | 0.0 | 4 | Staff |
| Overall | 02/29/20 | 12/07/21 | 166 | 56.6 | 737110 | 0.25 | 8610 | 1.3 | 1850 | |
Source: Own elaboration with data from: (OIE-WAHIS, 2021)
In addition to anorexia, fatigue, depression, and stooped posture (Chan et al., 2020; Munster et al., 2020), in farm animals, alterations in reproductive parameters, increased mortality, and development of secondary infections have been reported (Oreshkova et al., 2020).
The principal postmortem lesions described in animals (Table 4) show acute interstitial pneumonia with the presence of edema, hemorrhages, and areas of pulmonary consolidation; epithelial necrosis in the anterior airways; decrease in the alveolar lumen; infiltration of inflammatory and immune cells; and hematological alterations and radiographic changes in the pulmonary pattern (Boudewijns et al., 2020; Pruijssers et al., 2020; Schlottau et al., 2020).
Table 4 Clinical manifestations in animals infected with SARS-CoV-2
| Animal species | Clinical signs | Lesions and alterations | References |
|---|---|---|---|
| Domestic animals Cat | Ocular and nasal discharge, fever, dyspnea, sneezing, wheezing, hypothermia, lethargy, anorexy. | Inflamed and hemorrhagic nasal, tracheal and pulmonary mucosal epithelia. | (OIE-WAHIS, 2021; Oreshkova et al., 2020) |
| Dog | Dry cough, nasal discharge, fever, dyspnea, abnormal lung sounds, positive palm percussion, pharyngitis, bronchitis, polypnea, tachycardia, fatigue, anorexy, depression, lymph adenomegaly. | NR | (OIE-WAHIS, 2021) |
| Zoo animals Lion | Dry cough, serous nasal discharge, dyspnea. | NR | (OIE-WAHIS, 2021) |
| Snow leopard | Dry cough, nasal discharge, wheezing, sneezing. | NR | (OIE-WAHIS, 2021) |
| Tiger | Dry cough, nasal discharge, dyspnea, anorexy, neurological alterations. | Trachea and bronchi with presence of bloody mucus. | (OIE-WAHIS, 2021) |
| Farm animals Mink | Cough, nasal discharge, severe dyspnea, sneezing, Bodyweight loss, inappetence, increase in mortality, reproductive failure. | Diffuse interstitial pneumonia with hyperemia, loss of alveolar lumina, sepsis, edema, diffusely dark lung lobes, dystocia, clinical alterations compatible with chronic Aleutian disease. | (Abdel-Moneim & Abdelwhab, 2020; OIE-WAHIS, 2021; Sit et al., 2020) |
| Lab animals Monkey | Hyperthermia, Bodyweight loss. | Radiographic changes in lungs, macroscopic and inflammatory alterations in the lung, heart, liver and stomach, hematological alterations (lymphocytosis, monocytosis and increased cytokine activity). | (Munster et al., 2020) |
| Macaque | Cough, fever, changes in respiratory pattern, tachypnea, asthenia, bodyweight loss, anorexy, piloerection, hunched posture, pale appearance and dehydration. | Interstitial pneumonia, radiographic changes in lungs, edema, hemorrhage, lung congestion and consolidation, hematological alterations (leukocytosis). | (Woodby et al., 2020) |
| Ferret | Cough, fever, adynamic, anorexy. | Rhinitis with epithelial degeneration and necrosis, bronchiolitis, mixed cellular infiltrates in lungs, increase in alveolar macrophages, severe lymphoplasmacytic vasculitis and perivasculitis. | (Lu et al., 2020; Oreshkova et al., 2020) |
| Mice | Dyspnea, bodyweight loss, thin hair. | Pneumonia with infiltration of inflammatory and immune cells, peribronchiolar inflammation, hemorrhages in lungs and alveoli. | (Boudewijns et al., 2020) |
| Hamster | Tachypnea, lethargy, anorexy, piloerection, hunched posture. | Pneumonia, hemorrhages in lungs and alveoli, Lungs with presence of edema, hemorrhage and severe consolidation, multifocal necrotizing bronchiolitis and leukocyte infiltration. | (Lotfi & Rezaei, 2020; Schlottau et al., 2020) |
| Shrew | Fever. | Histopathological changes in liver, spleen, intestines, pancreas, kidney, heart, lung and brain. | (Pruijssers et al., 2020) |
Source: Own elaboration with data from: (Boudewijns et al., 2020; OIE-WAHIS, 2021; Pruijssers et al., 2020; Schlottau et al., 2020; Shi et al., 2020; Zhao et al., 2020)
Diagnostic techniques
Several diagnostic methods have been developed for clinical use or research. Their rationale is based mainly on the detection of nucleic acids or immunological and pathological changes. The WHO has established the importance of the use of SARS-CoV2 diagnostic tests that allow surveillance of the disease, limit its spread, assess the epidemiological risk, trace positive cases, local control of outbreaks, and determine previously infected individuals. COVID-19 tests are divided into two groups based on their diagnostic foundation (Table 5): tests for the detection of viral nucleic acid and tests for the detection of antigens or antibodies (Jarrom et al., 2020).
Table 5 Diagnostic techniques for SARS-CoV-2
| Test | Sample | Advantages | Disadvantages |
|---|---|---|---|
| Nucleic acid tests for viral RT-PCR | Nasopharyngeal or oropharyngeal swab, bronchoalveolar lavage, tracheal aspirates, saliva | High sensitivity, specificity, performance and reliable | Sensitivity may be affected by sampling errors or insufficient viral load |
| RT-LAMP | Blood | High sensitivity and specificity, easy to use | Complex primers, susceptible to amplification and false positives |
| RT-RPA | Blood | High sensitivity and specificity, easy to use | Complex primers, susceptible to amplification and false positives |
| CRISPR | Blood | High sensitivity and specificity, reliable, easy visual readability | Not tested for SARS-Cov-2. |
| Antigen or antibodies detection tests ELISA | Blood and saliva | Easy to use | Not as accurate as the RTPCR test, with false positives and negatives. |
| EIA | Nasopharyngeal swab and saliva | Easy to use, high performance and availability | Lack of knowledge and inability to confirm the antibodies |
| LFIA | Blood and saliva | Independent of laboratory equipment | Questionable sensitivity and specificity |
| IFT | Blood | No analyzer is required but an IF microscope is needed. | Low throughput, requires experience, discrimination of other coronavirus antibodies, time consuming |
| DB/WB | Blood | Discrimination of other coronavirus antibodies | Not common, WB experience required |
| VNT | Blood | Functional information | Biosafety level 3 lab required |
RT-PCR, real-time reverse transcriptase/polymerase chain reaction; RT-LAMP, reverse transcriptase loopmediated isothermal amplification; RT-RPA, reverse transcriptase recombinase polymerase amplification; CRISPR, clustered regularly interspaced short palindromic repeats; ELISA, enzyme-linked immunosorbent assay; EIA, enzyme immunoassay; LFIA, lateral flow immunoassay; SVNA, serum virus neutralization assay; IFT, immunofluorescence test; DB/WB, Dot blot/Western blot; VNT, virus neutralization test. Source: Own elaboration with data from from: (Abduljalil, 2020; D’Cruz et al., 2020; Gao & Quan, 2020; Hu et al., 2021; Özçürümez et al., 2020; Ravi et al., 2020).
Molecular tests are based on the presence of nucleic acids in a sample to make a diagnosis. These tests can detect nucleic acids prior transcription and are therefore considered essential for the diagnosis of diseases of viral origin. Some of these tests use procedures to identify and amplify viral nucleic acid, such as real-time reverse transcriptase PCR, reverse transcriptase loop-mediated isothermal amplification and reverse transcriptase recombinase polymerase amplification (Jarrom et al., 2020). These techniques have high sensitivity, specificity, and performance and are reliable and easy to implement. However, they have a disadvantage in their complex design of primers and alteration in sensitivity due to an insufficient viral load, in addition to not indicating whether the disease has been present previously (Gao & Quan, 2020; Jarrom et al., 2020; Li et al., 2020).
Serological tests focus on the detection of antigens or antibodies against SARS-CoV-2. These tests are easy to perform, have a high yield, and allow working many samples in a short time. However, they are less accurate and are likely to give false positives and negatives. Some of the techniques used on this basis are enzyme-linked immunosorbent assay (ELISA), enzyme immunoassay, lateral flow immunoassay, serum virus neutralization assay, immunofluorescence test, dot blot/western blot, and virus neutralization test. These techniques allow a diagnosis to be made in the laboratory or in an environment close to the patient, allowing the identification of sick, healthy, convalescent, reinfected, or previously infected animals (Gao & Quan, 2020; Jarrom et al., 2020).
ELISA detects the presence and concentration of immunoglobulin G or immunoglobulin M antibodies in the blood but have low specificity and cannot detect viral variables because the antibodies produced are very stable between species (Li et al., 2020; Michel et al., 2020; Sidiq et al., 2020). The western blot technique is based on the presence of viral or immune proteins in blood serum, and it can identify the molecular weight of unknown proteins other than a virus. However, it is a very laborious and time-consuming technique and is recommended more for research than for diagnosis (Sidiq et al., 2020). Techniques such as immunohistochemistry make it possible to demonstrate the damage caused by the virus in the various affected tissues and is used as a support for other diagnostic techniques. This technique provides physical evidence of the lesions caused by the virus in addition to being able to reveal its presence through high resolution microscopes (Gao & Quan, 2020; Nguyen et al., 2021).
Treatment
Therapeutic strategies to combat SARS-CoV-2 infection focus on reducing the severity of the disease because there are no specific drugs for treatment in animals. Even some of the most used drugs in humans (paracetamol and naproxen) are not recommended for use in dogs and cats (Papich, 2015); however, it has been recommended that the therapeutic protocol used in humans be adapted for animals according to the drugs approved for veterinary use (Ahn et al., 2020; Guo et al., 2020; Izda et al., 2020). Therapeutic indications have been established in dogs and cats with acute respiratory problems applicable to the alterations induced by SARS-CoV-2 (Nelson & Couto, 2019; Papich, 2015; Plumb, 2018). These are aimed at controlling each of the different clinical manifestations present in this disease (Table 6), reducing inflammation, edema and lack of pulmonary oxygenation through the use of drugs such as ibuprofen, acepromazine and oxygen administration respectively, as well as cough control (dextromethorphan, butorphanol) and bronchoconstriction (terbutaline, aminophylline). The use of antiviral drugs that inhibit viral replication processes (acyclovir, oseltamivir, ribavirin) is also suggested, as well as the administration of antibiotics that prevent the development of secondary infections of the respiratory tract that could be originated as a consequence of the alterations and lesions caused by SARS-CoV-2 (Ahn et al., 2020; Das et al., 2021; Nelson & Couto, 2019; Papich, 2015; Plumb, 2018).
Table 6 Suggested therapy for SARS-CoV-2-infected dogs and cats
| Therapeutic Indications | Optional drugs | Animal species/ Dosage | Administration via | Usual Interval (h/d) | |
|---|---|---|---|---|---|
| Dog | Cat | ||||
| Pain alleviation | Butorphanol (mg/kg) | 0.2-0.4 | 0.2-0.8 | IV, IM | 8-12/5 |
| Buprenorphine (mg/kg) | 0.005 | 0.005 | IV, IM | 4-8/3 | |
| Inflammation | Ibuprofen (mg/kg) | 5.0 | NR | PO | 12/5 |
| Prednisolone (mg/kg) | 0.5-1 | 0.5-1 | IV, IM, PO | 12-24/5 | |
| Pulmonary edema | Acepromazine (mg/kg) | 0.05 | 0.05 | IV, SC | 6-8/3 |
| Bronchospasm | Terbutaline (mg/kg) | 1.25-5 | 0.1 | PO | 8-12/5 |
| Aminophylline (mg/kg) | 11 | 5 | PO | 8-12/5 | |
| Oxtriphylline (mg/kg) | 14-47 | NR | PO | 8/5 | |
| Nonproductive cough | Dextromethorphan (mg/kg) | 1-2 | 0.5-2 | PO | 6-8/5 |
| Butorphanol (mg/kg) | 0.5 | NR | PO | 6-12/5 | |
| Hydrocodone (mg/kg) | 0.2-0.5 | NR | PO | 8-12/5 | |
| Nebulization with SSF+O2 (L/min) | 4-10 | 4-10 | IN | 4-12/5 | |
| Lower oxygen saturation Oxygen 50-60% (L/min) | 8-12 | 8-12 | Mask | Q.S. | |
| Oxygen 60% (L/min) | 2-3 | 2-3 | Oxygen cage | Q.S. | |
| Oxygen 100% (L/kg/min) | 0.2 | 0.2 | Endotracheal tube | Q.S. | |
| Secondary infections Azithromycin (mg/kg) | 5-10 | 5-10 | PO | 24/7 | |
| Viral sepsis Acyclovir (mg/kg) | 5-10 | 5-10 | PO | 6/10 | |
| Oseltamivir (mg/kg) | 2.2 | NR | PO | 12/7 | |
| Ribavirin (mg/kg) | 5-10 | 5.0-5-5 | PO | 12-24/7 | |
NR, Not reported. Source; IV, intravenous; IM, intramuscular; PO, per os; IN, intranasal; SC, subcutaneous; Q.S., quantum satis. Source: Own elaboration with data from: (Ahn et al., 2020; Das et al., 2021; Nelson & Couto, 2019; Papich, 2015; Plumb, 2018).
Control
The OIE has emitted a series of recommendations focused on the control of SARS-CoV2 in animals and in the workers in charge of their care and handling. These measures seek to minimize the spread of the disease and to avoid the appearance of viral mutations. However, the capacity to reduce the risk of the introduction and spread of SARS-CoV-2 in farm, domestic, and zoo animals varies greatly among countries because they apply different biosecurity, zoosanitary surveillance, and public health measures (OIE, 2021b).
To mitigate the risk of introduction and spread in fur farms and zoo animals, it is recommended to reinforce biosecurity measures in the facilities, to guarantee the use of personal protective equipment for workers and visitors, and to promote handwashing and disinfection after working with animals (OIE, 2021b). For workers who are in contact with animals, some of the control measures recommended by the OIE include respecting personal distance, avoiding large concentrations of people in common areas, not rotating workers between farms, informing workers about the pathways of spread of SARS-CoV2 in animals, and preventing workers with symptoms compatible with COVID-19 or living with someone with such symptoms from entering work facilities (OIE, 2021a).
In the case of an outbreak of SARS-CoV-2 in animals and workers, it is recommended to perform the sequencing, phylogenetic analysis, and comparison of genetic sequences of the viruses in all positive cases. Doing so allows the identification of mutations in the viral genome. Uninfected workers should inspect all animals for clinical respiratory or gastrointestinal signs, increase the level of personal protective equipment when handling sick or dead animals, immediately isolate animals positive in screening tests, and reduce the number of people interacting with these animals (OIE, 2021a).
Vaccines against SARS-CoV-2 in animals
The WHO has reported the importance of the use of vaccines in animals to protect against infection and prevent the spread of viral mutations to humans. To combat the possible threat of transmission from animals to humans and the increase in viral variants, an inactivated vaccine against COVID-19 called Karnivak-Kov or Carnivac-Cov was developed in Russia for use in carnivorous animals, and it is a safe, innocuous, highly immunogenic vaccine capable of producing immunity for at least six months after vaccination (Chavda et al., 2021). For the development of this vaccine, the SARS-CoV-2 virus was inactivated, causing the virus to be deficient in viral replication but with the capacity to be recognized by the host immune system and to provoke humoral and cellular immune responses against viral antigens (de Andrade et al., 2021). In the United States, the EvviVax laboratory conducted tests in felines vaccinated with DNA-Evvivax
LinearDNA™ producing neutralizing antibodies in 100% of the cases (Evvivax, 2021). Similarly, Zoetis Laboratories developed a vaccine against SARS-CoV-2 for great apes and ferrets and a new vaccine for zoo animals (Sharun et al., 2021; Zoetis, 2021).
Conclusion
The destruction of natural habitats, globalization, and the flow of travelers throughout the world contributed greatly to SARS-CoV-2, causing a pandemic in a relatively short time since December 2019, when it began to spread. On January 30, it was declared a worldwide emergency, and in a short time, the first cases occurred in America on April 6 and in Europe on April 28. As a result, protocols began to be applied worldwide to curb the pandemic, some with greater success than others, based on previous pandemics. The purpose of identifying the animal origin of SARS-CoV-2 and the way in which the virus is transmitted between different animal species is to establish strategies for a diagnostic and therapeutic approach and to establish control measures to help predict and prevent the future diffusion of the pandemic.
The various studies suggest that ferrets, minks, and felines are considered highly susceptible species to SARS-CoV-2, whereas dogs have a low susceptibility, and farm animals (cattle, pigs, and poultry) are not naturally susceptible. The above information suggests that domestic and zoo animals have become accidental hosts of SARS-CoV-2, becoming infected by contact with affected humans with whom them coexist. The concern remains that domestic and zoo animals could become a risk for maintaining mutant variants of the virus, which could eventually return to human populations.
REFERENCES
Abdel-Moneim AS, Abdelwhab EM. 2020. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 9(7):529. https://doi.org/10.3390/pathogens9070529 [ Links ]
Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, Kim BT, Kim SJ. 2020. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). https://doi.org/10.4014/jmb.2003.03011 [ Links ]
Anand KB, Karade S, Sen S, Gupta RM. 2020. SARS-CoV-2: Camazotz’s Curse. Medical Journal, Armed Forces India. 76(2):136. https://doi.org/10.1016/j.mjafi.2020.04.008 [ Links ]
Boudewijns R, Thibaut HJ, Kaptein SJF, Li R, Vergote V, Seldeslachts L, de Keyzer C, Bervoets L, Sharma S, van Weyenbergh J. 2020. STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. BioRxiv. https://doi.org/10.1101/2020.04.23.056838 [ Links ]
Bulut C, Kato Y. 2020. Epidemiology of COVID-19. Turkish Journal of Medical Sciences. 50(SI-1):563-570. https://doi.org/10.3906/sag-2004-172 [ Links ]
Chan JFW, Zhang AJ, Yuan S, Poon VKM, Chan CCS, Lee ACY, Chan W, Fan Z, Tsoi HW, Wen L. 2020. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clinical Infectious Diseases. 71(9):2428- 2446. https://doi.org/10.1093/cid/ciaa325 [ Links ]
Chavda VP, Feehan J, Apostolopoulos V. 2021. A Veterinary Vaccine for SARS-CoV-2: The First COVID-19 Vaccine for Animals. Vaccines. 9(6):631. https://doi.org/10.3390/vaccines9060631 [ Links ]
Chen Z, Boon SS, Wang MH, Chan RWY, Chan PKS. 2021. Genomic and evolutionary comparison between SARS-CoV-2 and other human coronaviruses. Journal of Virological Methods. 289:114032. https://doi.org/10.1016/j.jviromet.2020.114032 [ Links ]
Chowdhury SD, Oommen AM. 2020. Epidemiology of COVID-19. Journal of Digestive Endoscopy. 11(1):3. https://doi.org/10.1055/s-0040-1712187 [ Links ]
Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, Sagnelli C, Bianchi M, Bernardini S, Ciccozzi M. 2020. COVID-19 Outbreak: An Overview. Chemotherapy. 64(5-6): 215-223. https://doi.org/10.1159/000507423 [ Links ]
Das A, Roy S, Swarnakar S, Chatterjee N. 2021. Understanding the immunological aspects of SARS-CoV-2 causing COVID-19 pandemic: A therapeutic approach. Clinical Immunology. 108804. https://doi.org/10.1016/j.clim.2021.108804 [ Links ]
D’cruz RJ, Currier AW, Sampson VB. 2020. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Frontiers in Cell and Developmental Biology. 8:468. https://doi.org/10.3389/fcell.2020.00468 [ Links ]
De Andrade JF, de Lima Cruz IR, de Sampaio FMS, da Silva CGL, Lopes MR, Gadelha MSV. 2021. Pesquisa de SARS-COV-2 em cães e gatos: relatos de casos na literatura. Brazilian Journal of Development: 7(5):45198-45209. https://doi.org/10.34117/bjdv7n5101 [ Links ]
Evvivax. 2021. December 30). Evvivax and Applied DNA Veterinary COVID-19 Vaccine Candidate Induces Neutralizing Antibodies in 100% of Trial Cohort Against B.1.1.7, P1, and B.1.526 Variants. https://www.Evvivax.Com/News.Html [ Links ]
Fasina FO. 2020. Novel coronavirus (2019-nCoV) update: What we know and what is unknown. Asian Pacific Journal of Tropical Medicine. 13(3):97. https://doi.org/10.4103/1995-7645.277795 [ Links ]
Gao J, Quan L. 2020. Current Status of Diagnostic Testing for SARS-CoV-2 Infection and Future Developments: A Review. Medical Science Monitor. International Medical Journal of Experimental and Clinical Research. 26:e928552-1. https://doi.org/10.12659/MSM.928552 [ Links ]
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. 2020. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Military Medical Research. 7(1):1-10. https://doi.org/10.1186/s40779-020-00240-0 [ Links ]
Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, Ehrlich M, Grieser J, Winston J, Lombardi D. 2022. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 602(7897):481-486. https://doi.org/10.1038/s41586-021-04353-x [ Links ]
HU B, Guo H, Zhou P, Shi ZL. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology. 19(3):141-154. https://doi.org/10.1038/s41579-02000459-7 [ Links ]
ICTV (International Committee on Taxonomy of Viruses). 2021. https://talk.ictvonline.org/ [ Links ]
Izda V, Jeffries MA, Sawalha AH. 2020. COVID-19: A review of therapeutic strategies and vaccine candidates. Clinical Immunology. 222:108634. https://doi.org/10.1016/j.clim.2020.108634 [ Links ]
Jalava K. 2020. First respiratory transmitted food borne outbreak?. International Journal of Hygiene and Environmental Health. 226:113490. https://doi.org/10.1016/j.ijheh.2020.113490 [ Links ]
Jarrom D, Elston L, Washington J, Prettyjohns M, Cann K, Myles S, Groves P. 2020. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ EvidenceBased Medicine. 27(1):33-45. https://doi.org/10.1136/bmjebm-2020-111511 [ Links ]
Ji W, Li X. 2020. Response to “Comments on" Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross/species transmission from snake to human” and “Codon bias analysis alone is uninformative for identifying host (s) of new. Journal of Medical Virology. https://doi.org/10.1002/jmv.26048 [ Links ]
Ji W, Wang W, Zhao X, Zai J, Li X. 2020a. Cross/species transmission of the newly identified coronavirus 2019/nCoV. Journal of Medical Virology. 92(4):433-440. https://doi.org/10.1002/jmv.25682 [ Links ]
Ji W, Wang W, Zhao X, Zai J, Li X. 2020b. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. Journal of Medical Virology. 92(4):433-440. https://doi.org/10.1002/fut.22099 [ Links ]
Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. 2020. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases. 26(7):1542. https://doi.org/10.3201/eid2607.200092 [ Links ]
Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W. 2020. Development and clinical application of a rapid IgM/IgG combined antibody test for SARS/CoV/2 infection diagnosis. Journal of Medical Virology. 92(9):1518-1524. https://doi.org/10.1002/jmv.25727 [ Links ]
Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J. 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)?. PLoS Pathogens. 16(5):e1008421. https://doi.org/10.1371/journal.ppat.1008421 [ Links ]
Lotfi M, Rezaei N. 2020. SARS/CoV/2: a comprehensive review from pathogenicity of the virus to clinical consequences. Journal of Medical Virology. 92(10):1864-1874. https://doi.org/0000-0002-3836-1827 [ Links ]
Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, Kuang D, Yang M, Yang J, Ma C. 2020. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. BioRxiv. https://doi.org/10.1101/2020.04.08.031807 [ Links ]
Majumder J, Minko T. 2021. Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19. The AAPS Journal. 23(1):1-22. https://doi.org/10.1208/s12248-020-00532-2 [ Links ]
Michel M, Bouam A, Edouard S, Fenollar F, di Pinto F, Mège J, Drancourt M, Vitte J. 2020. Evaluating ELISA, immunofluorescence, and lateral flow assay for SARS-CoV-2 serologic assays. Frontiers in Microbiology. 11:597529. https://doi.org/10.3389/fmicb.2020.597529 [ Links ]
Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B. 2020. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 585(7824):268-272. https://doi.org/10.1038/s41586-020-2324-7 [ Links ]
Nelson RW, Couto CG. 2019. Small Animal Internal Medicine-E-Book. Elsevier Health Sciences. ISBN 978-0-323-57014-5 [ Links ]
Nguyen, D, Skelly D, Goonawardane N. 2021. A Novel Immunofluorescence Assay for the Rapid Serological Detection of SARS-CoV-2 Infection. Viruses. 13(5):747. https://doi.org/10.3390/v13050747 [ Links ]
Oie, W. O. for A. H. (World Organisation for Animal Health). 2021a. Guidance on working with farmed animals of species susceptible to infection with SARS-CoV-2. https://www.oie.int/fileadmin/Home/MM/Draft_OIE_Guidance_farmed_animals_cleanMS05.11.pdf [ Links ]
Oie, W. O. for A. H. (World Organization for Animal Health). 2021b. SARS-CoV-2 in animals used for fur farming: GLEWS+ risk assessment, 20 January 2021. World Health Organization. https://www.who.int/publications/i/item/WHO-2019-nCoV-fur-farming-riskassessment-2021.1 [ Links ]
OIE-WAHIS. (World Organization for Animal Health). 2021. World Animal Health Information System. https://wahis.oie.int/#/events [ Links ]
Oreshkova N, Molenaar RJ, Vreman S, Harders F, Munnink BBO, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS. 2020. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. 25(23):2001005. https://doi.org/10.2807/1560-7917.ES.2020.25.23.2001005 [ Links ]
Özçürümez MK, Ambrosch A, Frey O, Haselmann V, Holdenrieder S, Kiehntopf M, Neumaier M, Walter M, Wenzel F, Wölfel R. 2020. SARS-CoV-2 antibody testing- questions to be asked. Journal of Allergy and Clinical Immunology. 146(1):35-43. https://doi.org/10.1016/j.jaci.2020.05.020 [ Links ]
Papich MG. 2015. Saunders handbook of veterinary drugs-e-book: small and large animal. Elsevier Health Sciences. ISBN 978-0-323-24485-5 [ Links ]
Plumb DC. 2018. Plumb’s Veterinary Drug Handbook: Desk. John Wiley & Sons. ISBN 978-1-1193-4445-2 [ Links ]
Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, Dinnon III KH, Yount BL, Agostini ML, Stevens LJ, Chappell JD. 2020. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Reports. 32(3):107940. https://doi.org/10.1016/j.celrep.2020.107940 [ Links ]
Ravi N, Cortade DL, Ng E, Wang SX. 2020. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosensors and Bioelectronics, 165:112454. https://doi.org/10.1016/j.bios.2020.112454 [ Links ]
Rothan HA, Byrareddy SN. 2020. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. Journal of Autoimmunity. 102433. https://doi.org/10.1016/j.jaut.2020.102433 [ Links ]
Ruiz-Arrondo I, Portillo A, Palomar AM, Santibanez S, Santibanez P, Cervera C, Oteo JA. 2020. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV2 in Europe. Transboundary and emerging diseases. 68(2):973-976. https://doi.org/10.1101/2020.05.14.20101444 [ Links ]
Salata C, Calistri A, Parolin C, Palù G. 2019. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathogens and Disease. 77(9):ftaa006. https://doi.org/10.1093/femspd/ftaa006 [ Links ]
Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T. 2020. Experimental transmission studies of SARS-CoV-2 in fruit bats, ferrets, pigs and chickens. Lancet Microbe. 1(5):e218-e225. https://doi.org/10.1016/S2666-5247(20)30089-6 [ Links ]
Sharun K, Dhama K, Pawde AM, Gortázar C, Tiwari R, Bonilla-Aldana DK, RodriguezMorales AJ, de la Fuente J, Michalak I, Attia YA. 2021. SARS-CoV-2 in animals: potential for unknown reservoir hosts and public health implications. Veterinary Quarterly. 41(1):181-201. https://doi.org/10.1080/01652176.2021.1921311 [ Links ]
Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. 2020. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 24(3):91-98. https://doi.org/10.1016/j.jare.2020.03.005 [ Links ]
Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 368(6494):1016-1020. https://doi.org/10.1126/science.abb7015 [ Links ]
Sidiq Z, Hanif M, KumarDwivedi K, Chopra KK. 2020. Benefits and limitations of serological assays in COVID-19 infection. Indian Journal of Tuberculosis. 67(4):S163S166. https://doi.org/10.1016/j.ijtb.2020.07.034 [ Links ]
Singhal T. 2020. A review of coronavirus disease-2019 (COVID-19). The Indian Journal of Pediatrics. 1-6. https://doi.org/10.1007/s12098-020-03263-6 [ Links ]
Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW. 2020. Infection of dogs with SARS-CoV-2. Nature. 586(7831):776-778. https://doi.org/10.1038/s41586-020-2334-5 [ Links ]
Suarez DL, Pantin-Jackwood MJ, Swayne DE, Lee SA, Deblois SM, Spackman E. 2020. Lack of susceptibility of poultry to SARS-CoV-2 and MERS-CoV. BioRxiv. https://doi.org/10.1101/2020.06.16.154658 [ Links ]
Sun J, He WT, Wang L, Lai A, Ji X, Zhai X, Li G, Suchard MA, Tian J, Zhou J. 2020. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends in Molecular Medicine. 26(5):483-495. https://doi.org/10.1016/j.molmed.2020.02.008 [ Links ]
Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z. 2020. On the origin and continuing evolution of SARS-CoV-2. National Science Review. 7(6):1012-1023. https://doi.org/10.1093/nsr/nwaa036 [ Links ]
Tiwari R, Dhama K, Sharun K, Iqbal Yatoo M, Malik YS, Singh R, Michalak I, Sah R, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. COVID-19: animals, veterinary and zoonotic links. Veterinary Quarterly. 40(1):169-182. https://doi.org/10.1080/01652176.2020.1766725 [ Links ]
WHO. (World Health Organization). (2020). COVID 19 Public Health Emergency of International Concern (PHEIC). Global research and innovation forum: towards a research roadmap. https://www.who.int/publications/m/item/covid-19-public-health-emergency-ofinternational-concern-(pheic)-global-research-and-innovation-forum [ Links ]
WHO. (World Health Organization). 2021. WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ [ Links ]
Wong G, Bi YH, Wang QH, Chen XW, Zhang ZG, Yao YG. 2020. Zoonotic origins of human coronavirus 2019 (HCoV-19/SARS-CoV-2): why is this work important?. Zoological Research. 41(3): 213. https://doi.org/10.24272/j.issn.2095-8137.2020.031 [ Links ]
Woodby B, Arnold MM, Valacchi G. 2020. SARS/CoV/2 infection, COVID/19 pathogenesis, and exposure to air pollution: What is the connection?. Annals of the New York Academy of Sciences. 1486(1):15-38. https://doi.org/10.1111/nyas.14512 [ Links ]
Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2020. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nature Structural & Molecular Biology. 27(8):763-767. https://doi.org/10.1038/s41594-020-0468-7 [ Links ]
Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X. 2020. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 583(7815):286-289. https://doi.org/10.1038/s41586-020-2313-x [ Links ]
Xie E, Guangzhou GRI. 2020. Why wild animals are a key ingredient in China’s coronavirus outbreak. Australasian Policing. 12(2):8. https://search.informit.org/doi/10.3316/informit.217111399726998 [ Links ]
Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. 2020. Zoonotic origins of human coronaviruses. International Journal of Biological Sciences. 16(10):1686. https://doi.org/10.7150/ijbs.45472 [ Links ]
Zhang T, Wu Q, Zhang Z. 2020. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology. 30(7):1346-1351.e2. https://doi.org/10.1016/j.cub.2020.03.022 [ Links ]
Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, Zhao S, Li J, Long H, Ding K. 2020. Susceptibility of tree shrew to SARS-CoV-2 infection. Scientific Reports. 10(1):1-9. https://doi.org/10.1038/s41598-020-72563-ww [ Links ]
Zheng J. 2020. SARS-CoV-2: an emerging coronavirus that causes a global threat. International Journal of Biological Sciences. 16(10):1678-1685. https://doi.org/10.7150/ijbs.45053 [ Links ]
Zoetis. 2021. Zoetis’ Emerging Infectious Disease Capabilities Support COVID-19 Solutions for Great Apes and Minks [Commercial]. https://www.zoetis.com/news-andmedia/feature-stories/posts/zoetis-emerging-infectious-disease-capabilities-supportcovid-19-solutions-for-great-apes-and-minks.aspx [ Links ]
Received: June 30, 2022; Accepted: May 30, 2023











 text in
text in 



