Durinskia baltica (Levander) Carty & Cox has been identified in Mexico as a phytoplanktonic species in Xochimilco Lake (Lira et al. 2017). It belongs to the family Kryptoperidiniaceae, and it is a dinotom, i.e., a dinophyte possessing a tertiary diatom-like endosymbiont. To confirm the identity of the Mexican material, we compared it with the original description of the genus and the iconotype of D. baltica, which relied on a population from a drainage ditch in College Station, Texas, USA (Carty & Cox 1986, Figures 11-14, p. 202). The name Durinskia baltica is considered a heterotypic synonym of Durinskia dybowskii (Wolozinska) Carty (Molinari-Novoa 2022); for other authors and with plausible arguments, the legitimacy of the name D. dybowskii is in doubt (INA 2023, Lira et al. 2022). This controversy is one of the reasons why in this work, we maintain the name D. baltica.
Lira & Tavera (2019) provided life cycle knowledge for this species. This knowledge made evident this organism's particular developmental strategies in the freshwater environment, which explains its dominance in the phytoplankton of Xochimilco Lake (Lira et al. 2017). Throughout its morphological study and isolation in culture, it was necessary to analyze the controversial systematic history of the genus, which is related to an extensive ecological record (Lira et al. 2022).
Durinskia baltica has been observed in freshwater, marine, and brackish waters, and its geographic distribution covers most continents (Kretschmann et al. 2018). Such a wide geographic and ecological distribution is difficult to explain for a single species. In Durinskia baltica, when analyzing the ecological significance of its presence in such different environments, apparently worldwide, it is necessary to face the historical complexity of its systematic circumscription. This issue has been extensively addressed in previous studies (Lira et al. 2022). There is also a description of Durinskia oculata (Stein) Hansen & Flaim as a distinct freshwater entity, recently confirmed with a genetic study of its type material (Kretschmann et al. 2018). To complete a comprehensive characterization of the population recorded in Mexico, its genetic analysis and molecular phylogenetic comparison with D. oculata and other freshwater species of the genus for which sequences exist in databases was mandatory. This analysis also included brackish water and marine sequences available for dinotoms. We used the 18S rDNA sector to amplify the sequence of the dinophyte, and as it is a dinotom, we also considered the genetic analysis of the endosymbiont based on the sector coding for the large subunit of ribulose, 1-5-bisphosphatase carboxylase/oxygenase (rbcL). In parallel and based on database sequences, we explored the rbcL sector in a Bayesian inference of present-day diatoms associated with dinotom endosymbionts.
The temporal component that places events in a phylogeny plays an essential role in understanding relationships among dinophytes that have colonized freshwater, as is the case for the species of our interest (Žerdoner-Čalasan et al. 2018, 2019). Even with the extent of the recent literature spectrum (Table S1, Supplementary material), it is notable that calibrated molecular dating is scarcely addressed for the family Kryptoperidiniaceae. Thus, we aimed to base the phylogenetic analysis on the fossil record and a relaxed molecular clock. In the evolutionary context of the colonization of freshwater environments by dinophytes, the case of the Mexican population stood out because the molecular calibration for the host sequence coincides with the geological origin proposed for the Xochimilco basin, from which the specimens studied here originate (Arce et al. 2019).
Material and methods
Study area. Durinskia material was based on isolated populations from Xochimilco Lake from 2010 to 2019 (Figure 1); this lake is currently a system of natural freshwater canals, sometimes very wide, located in central Mexico (19° 16' N, 99° 00' W; 19° 16' N, 99° 04' W; 19° 16' N, 99° 05' W). Observations of the organisms were made using differential interference contrast (DIC) with a Nikon-Optiphot photon microscope (Tokyo, Japan) equipped with a Nikon Coolpix S10 camera.
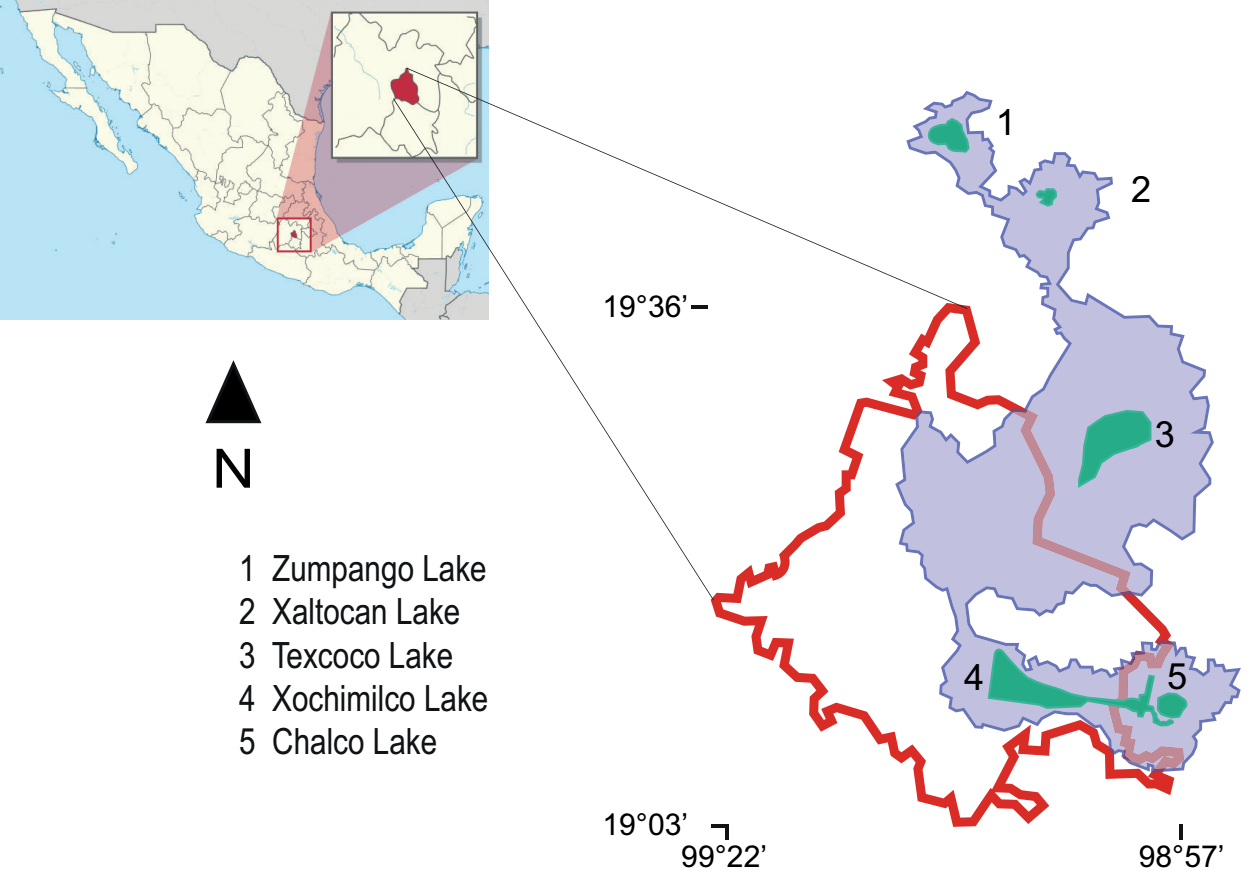
Figure 1 Map of Mexico showing (left) Mexico City (CDMX), filled in red. On the right, CDMX is bounded by a red line. The blue area delimits the original lake area (until the 12th century). Current lakes are shown in green. The number corresponds to the list of names. Map source: Commons license under GNU General Public https://commons.wikimedia.org/w/index.php?curid=1192145 (public domain).
Processing for scanning electron microscopy (Jeol 5410 LV microscope, Massachusetts, USA) followed standard protocols previously described in Lira et al. (2017). Dinokaryon observations were performed by staining single cells with 1 μl ml-1 of 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) (Sigma-Aldrich™ D95242, Missouri, USA). Epifluorescence image was acquired using an upright confocal microscope (TCS-SP8, LEICA, Wetzlar, Germany). Cells were excited with a 405 nm laser and emission was collected between 410-480 nm with a hybrid detector. Samples were observed with 63x/1.40 oil immersion objective.
Molecular phylogenies. We extracted genomic DNA from cultured cells using the Power Biofilm DNA Isolation Kit (MoBio Laboratories, California, USA) according to the manufacturer's specifications. For the amplification of the 18S rDNA region, four sets of primers were used (Tavera & Díez 2009, Lira 2018), and for the sector coding rbcL, we used specific diatom primers according to Tamura et al. (2005) (Table 1), following the protocols proposed by DNA polymerase Taq Master Mix (Ampliqon, Odense, Denmark) for both regions. For 18S rDNA, PCR amplification conditions were as follows: 95 °C / 15 min, followed by 40 cycles at 95 °C / 30 s; 60 °C / 30 s; 72 °C / one min, and a final extension at 72 °C / five min, to end with four °C / ∞. For rbcL, the conditions were: initial temperature of 94 °C / three min, followed by 35 cycles of 94 °C / one min; 50 °C / one min; 72 °C / one min, 72 °C / five min, and four °C / ∞.
Table 1 A) Primer sequences used to amplify the 18S rRNA and rbcL regions (specific for diatoms). B) Obtained sequences and size.
| A | ||
|---|---|---|
| Gene | Primer | Sequence |
| 18S rRNA | DbalF | 5'-AACCTGGTTGATCCYGCCAG-3' |
| 18S rRNA | DbalR | 5'-TGATCCTTCTGCAGGTTCACCTAC-3' |
| 18S rRNA | EUKA | 5'-AACCTGGTTGATCCTGCCAGT-3’ |
| 18S rRNA | EUKB | 5'-TGATCCTTCTGCAGGTTCACCTAC-3' |
| rbcL | DiatrbcL2F | 5'-ACAGTAAAACCTAAATTAGG-3' |
| rbcL | DiatrbcL5R | 5'-ATTTGACCACAGTGGATACC-3' |
| B | ||
| Obtained sequence | Base pairs | |
| EUKA (18S) | 1014 | |
| EUKB (18S) | 1223 | |
| DbalF (18S) | 600 | |
| DbalR (18S) | 600 | |
| CONTIG EUKA-EUKB (18S) | 1165 | |
| DiatrbcL2F (rbcL) | 407 | |
| DiatrbcL5R (rbcL) | 659 | |
| DiatrbcL2F- DiatrbcL5R (rbcL) | 560 |
Sequences (Sanger) were obtained through Macrogen Sequencing Services (Macrogen Inc. Korea) and manually cleaned and edited using the BioEdit editor (Hall 1999). Multiple sequence alignment methods were SINA for 18S rRNA sequences and MUSCLE for rbcL sequences available in the GenBank database (Edgar 2004, Pruesse et al. 2012, Sayers et al. 2022). Forty-one partial sequences of 1,300 bp, including the one in our study, supported the alignment of the 18S rRNA and 30 partial sequences of 1,400 bp supported the alignment of the rbcL gene. BEAST v. 2.6.7 software was the frame for Bayesian inference analysis of sequences and estimation of divergence times (Alekseyenko et al. 2022). The analysis in BEAST focused on a relaxed molecular clock with a log-normal distribution for interbranch rate variation and an assumption of independent rates between branches (gamma). In addition, we selected the GTR model of sequence evolution and a Yule-calibrated prior on evolutionary rates. Divergence dates were obtained from the dinophyte literature and from the fossil record calculated in the evolutionary Timetree of life, founded on diversification studies with multigene molecular clocks (Fensome et al. 1996, Berney & Pawlowski 2006, Parfrey et al. 2011, Heled & Drummond 2012, Kumar et al. 2017, Žerdoner-Čalasan et al. 2019). A series of 6'000,000 generations, with a 10 % burn-in, was completed on the BEAST dataset. Trees from BEAST analyses were summarized with Tree Annotator v. 1.7.1 and visualized with FigTree v. 1.4.4 (Drummond & Rambaut 2007, Rambaut 2018).
Results
Morphology. In the Mexican material, vegetative cells were spherical to ellipsoidal. Dorsoventrally, cells were slightly flattened, with 24.0-34.0 μm long dimensions and 18.0-22.0 μm wide. Generally, the epitheca and hypotheca were similar in size. The cingulum presented a downward planozone orientation, with a wide groove not reaching the antapex. A prominent red stigma locates within the hypotheca; it is hook-shaped, according to Lira et al. (2017). The epitheca presented the typical red accumulation bodies. The cell presented numerous golden, discoid chloroplasts scattered throughout (Figure 2A). The dinokaryon (nucleus) was located in the center of the cell and presented the condensed chromosomes characteristic of dinophytes; its dimensions were 16.0-24.0 μm long and 10.0-12.0 μm wide (Figure 2A, B). The thecal formula consisted of the following arrangement: Po, x, 4', 2a, 6'''', 5c, 4s, 5'''', 2''''' (Figures 3, 4). Thecal plates were asymmetrical. Plate 1' was rhomboidal and exceptionally large (Figure 3A, B). The plates showed fine, scattered pores and pores parallel to the suture lines (Figure 4A). The cell presented an inconspicuous ridge surrounding the apical pore (Figure 4B) and presented two intercalary plates located just above the 3" and 4" plates, one followed by the other: plate 1a, was smaller than plate 2a and diamond-shaped (Figures 3B, 4B). Through transmission electron microscopy, Lira et al. (2017) showed that the nucleus of endosymbiotic origin, associated with chloroplasts, is surrounded by a simple membrane, thus isolating it from the dinokaryon and host cytoplasm.
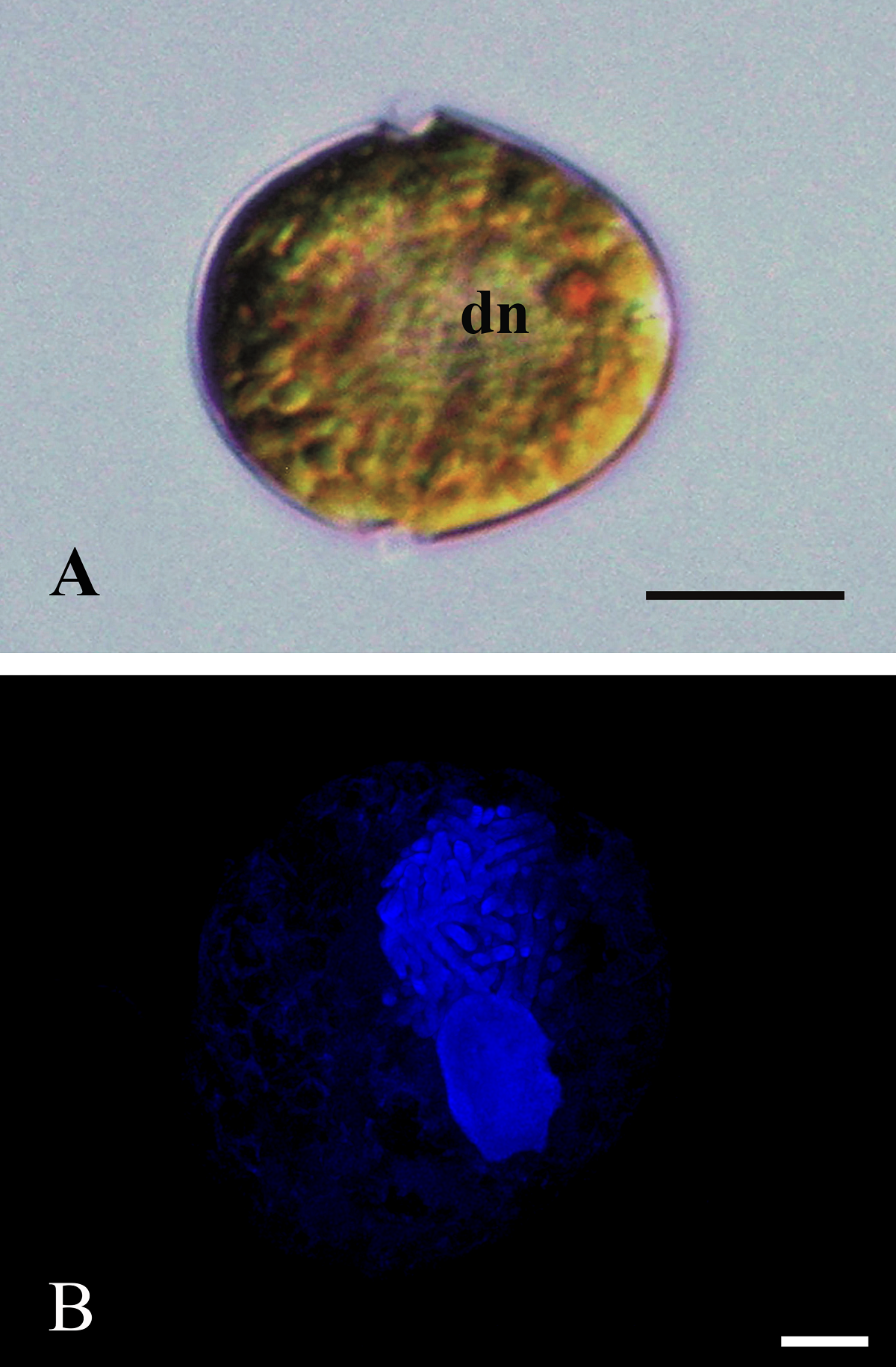
Figure 2 A): ML photograph of a D. baltica cell (sample from Xochimilco Lake) showing the typical dinophyte dinocaryon (dn) with condensed chromosomes and a red accumulation body in the epitheca; some chloroplasts towards the periphery of the cell. Bar = 10 μm. B): Epifluorescence photograph highlighting with DAPI staining the nucleus from D. baltica, and the nucleus from its endosymbiont (sample from Lago Xochimilco). Condensed chromosomes stand out in dinokaryon, besides the smaller endosymbiont nucleus. Bar = 5 μm.
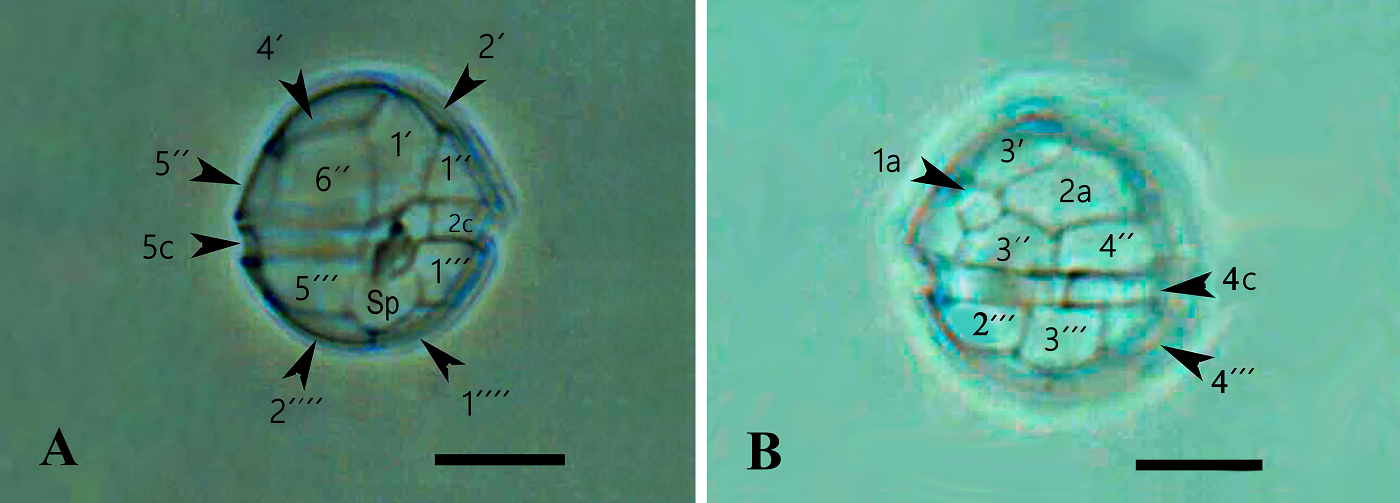
Figure 3 A): ML photograph of a D. baltica theca (sample from Xochimilco Lake) showing thecal plate nomenclature in ventral view. Bar = 10 μm. The posterior sulcal plate (Sp) reaches the antapical region just between the suture of antapical plates 1'''' and 2''''. Over Sp is visible the folding of the left sulcal plate (Sd). B): ML photograph of a D. baltica theca (sample from Xochimilco Lake) showing thecal plate nomenclature in dorsal view. Visible the two intercalary plates, 1a and 2a that characterize Durinskia species. Bar = 10 μm.
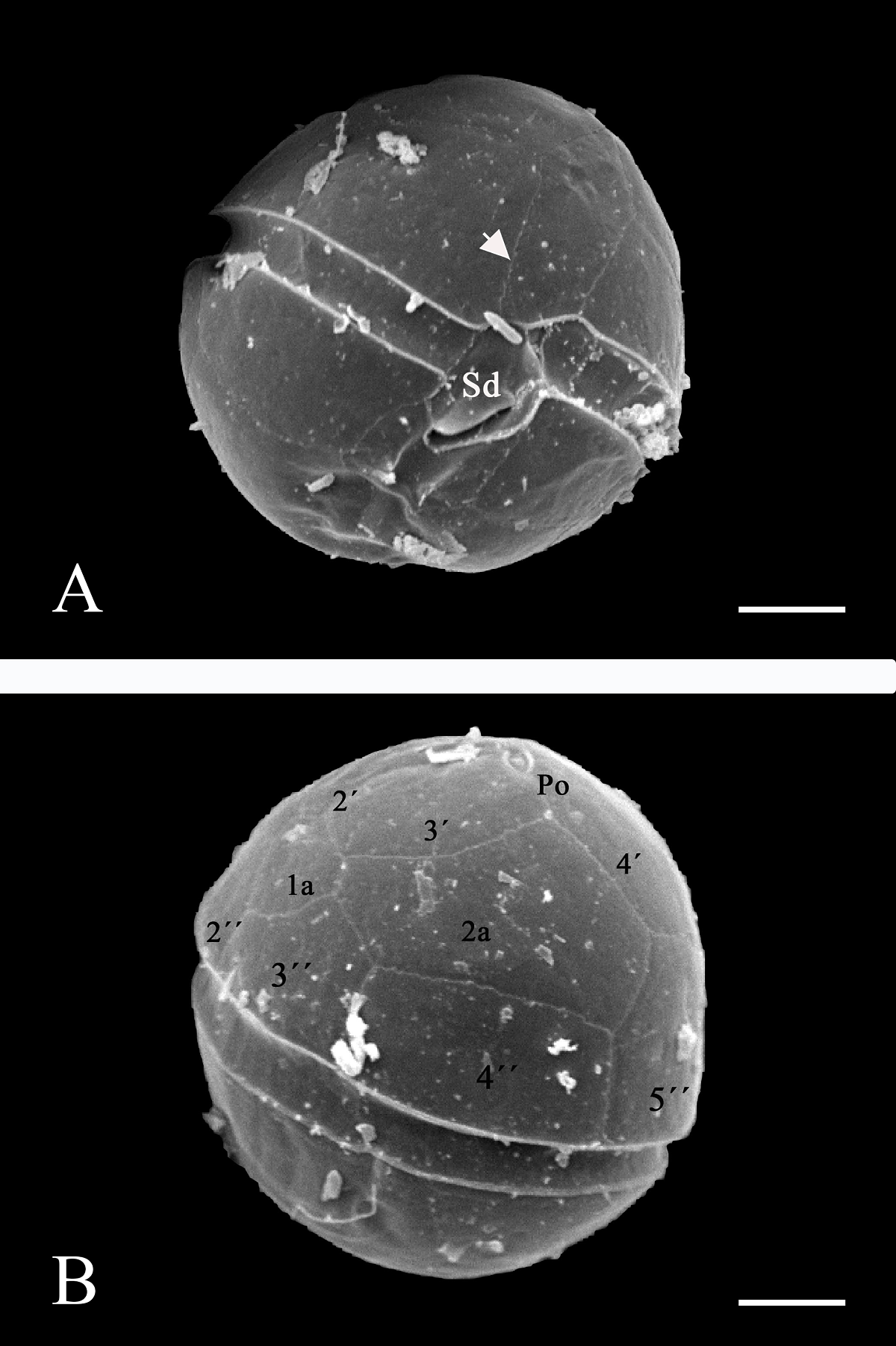
Figure 4 A): In ventral view, a SEM micrograph with backscattered electrons for 10 kV of a D. baltica theca (sample from Lago Xochimilco). The cingulum shows the Sd plate forming a fold over the sulcal area, the arrow points to the pores parallel to the suture lines. Bar = 5 μm. B): SEM micrograph with backscattered electrons for 10 kV of a D. baltica theca (sample from Lago Xochimilco) in dorsal view, with plate nomenclature in the epitheca. The apical pore (Po) and plate 1a, characteristic of the genus, stand out. Bar = 5 μm.
Genetic analysis. 18S rRNA sequences of populations isolated from cultures of Durinskia, selected from Xochimilco, Mexico, were examined using GenBank datasets for the same marker in this and other genera from several families of Dinophyta (41 sequences). This dataset was analyzed using Bayesian criteria. The monophyly of the family Kryptoperidiniaceae revealed a high support value (BPP 0.94) (Figure 5). Peridinium quinquecorne Abé was basal and formed the most senior group together with Perkinsus marinus (Mackin, Owen & Colliere) Levine, representing Alveolates (outgroup), which designated P. quinquecorne (circa 750 mybp) as one of the earliest dinophytes to diverge from Alveolata (about 1300 mybp, Parfrey et al. 2011). Diversifying in the Mesozoic era distinguished the Dinophyta group, probably initiated in the late Permian (Fensome et al. 1996, Žerdoner-Čalasan et al. 2019). Our results indicated that the ancestors of marine, brackish, and freshwater dinophytes appeared around the Ordovician-Silurian boundary (443.8 mybp). In the case of Durinskia, freshwater species were grouped with brackish-water species, sharing a common ancestor. The origin of these species was among the most recent. In the marine sequences, only that representing dinotoms with temporary endosymbionts such as D. kwazulunatalensis Yamada, Sim & Horiguchi and D. capensis Pienaar, Sakai & Horiguchi (Yamada et al. 2017, 2019). showed a recent origin (Figure 5).
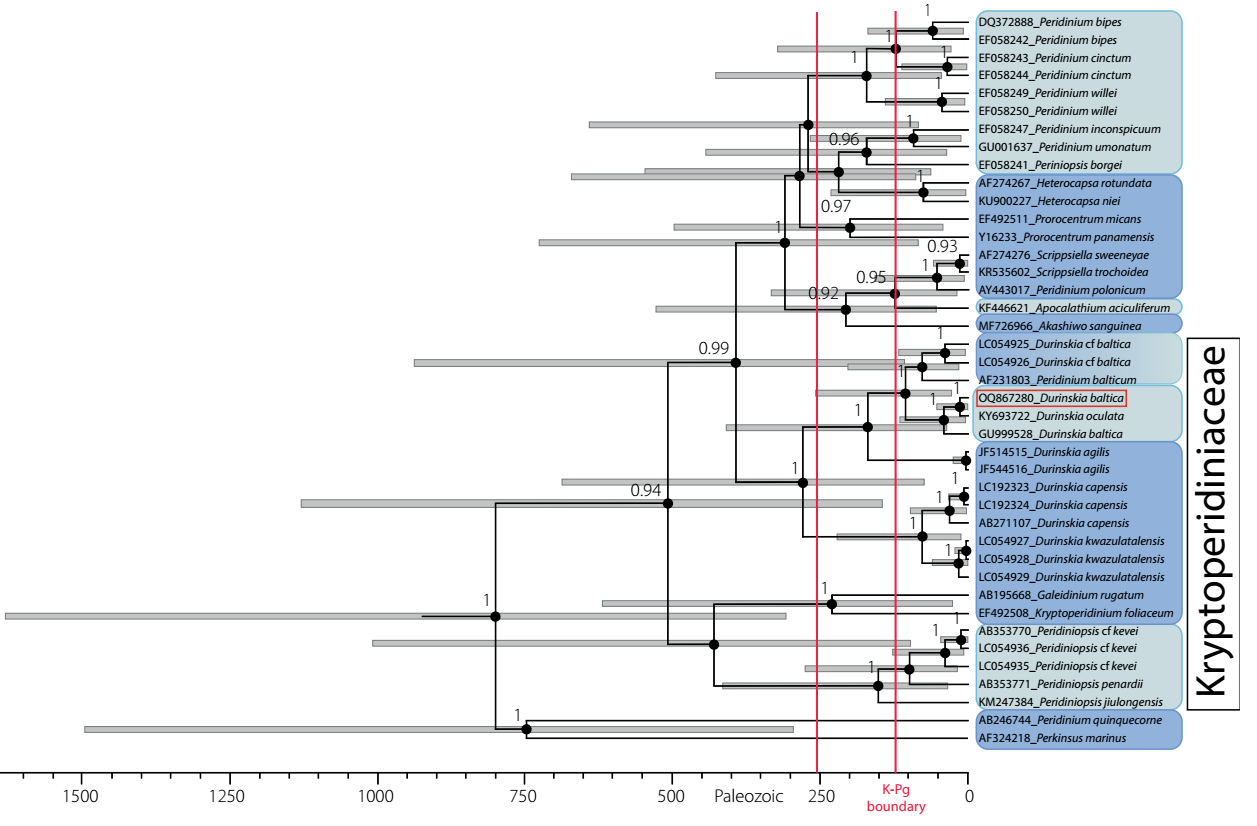
Figure 5 Dated phylogeny of Dinophyta obtained from BEAST analysis. Numbers at each node correspond to Bayesian posterior probability values. Gray bars represent 95 % confidence intervals for the height posterior density (HPD) in the estimates of divergence dates. The time scale is in millions of years before the present (mybp). Bayesian posterior probabilities < 90 are not shown, as the ages they represent are imprecise. The Mesozoic era is represented between the red vertical lines, indicating the K-Pg boundary. Light blue distinguishes freshwater taxa, dark blue marine taxa, and degraded blue brackish taxa. Durinskia baltica from Mexico framed in red.
In our results, several freshwater species in Kryptoperidiniaceae appear to have originated less than 50 mybp ago (Figure 5). The freshwater Durinskia species, Durinskia oculata (Europe) and Durinskia baltica (Mexico), originated 20 mybp. The origin calculated for the Mexico sequence was very close to the geological origin of the lake basin (Arce et al. 2019).
Thirty chloroplast sequences were analyzed with Bayesian criteria; rbcL was the marker used for the Mexican population and a GenBank dataset for the same marker in dinotoms and extant diatoms (Figure 6). The genetic analysis determined that the rbcL sequence from Xochimilco distinguished a tertiary endosymbiont of the Nitzschia type. Most of the endosymbiont sequences were associated with freshwater Bacillariophyceae diatom sequences, mainly with Nitzschia Hassall species, regardless of whether or not the endosymbiont corresponded to a brackish, marine, or freshwater host. Only the endosymbionts of Peridinium quinquecorne and Peridiniopsis Lemmermann species corresponded environmentally with freshwater or marine diatom sequences of the class Mediophyceae. In no case was the sequence of an endosymbiont associated with that of another endosymbiont from different host species.
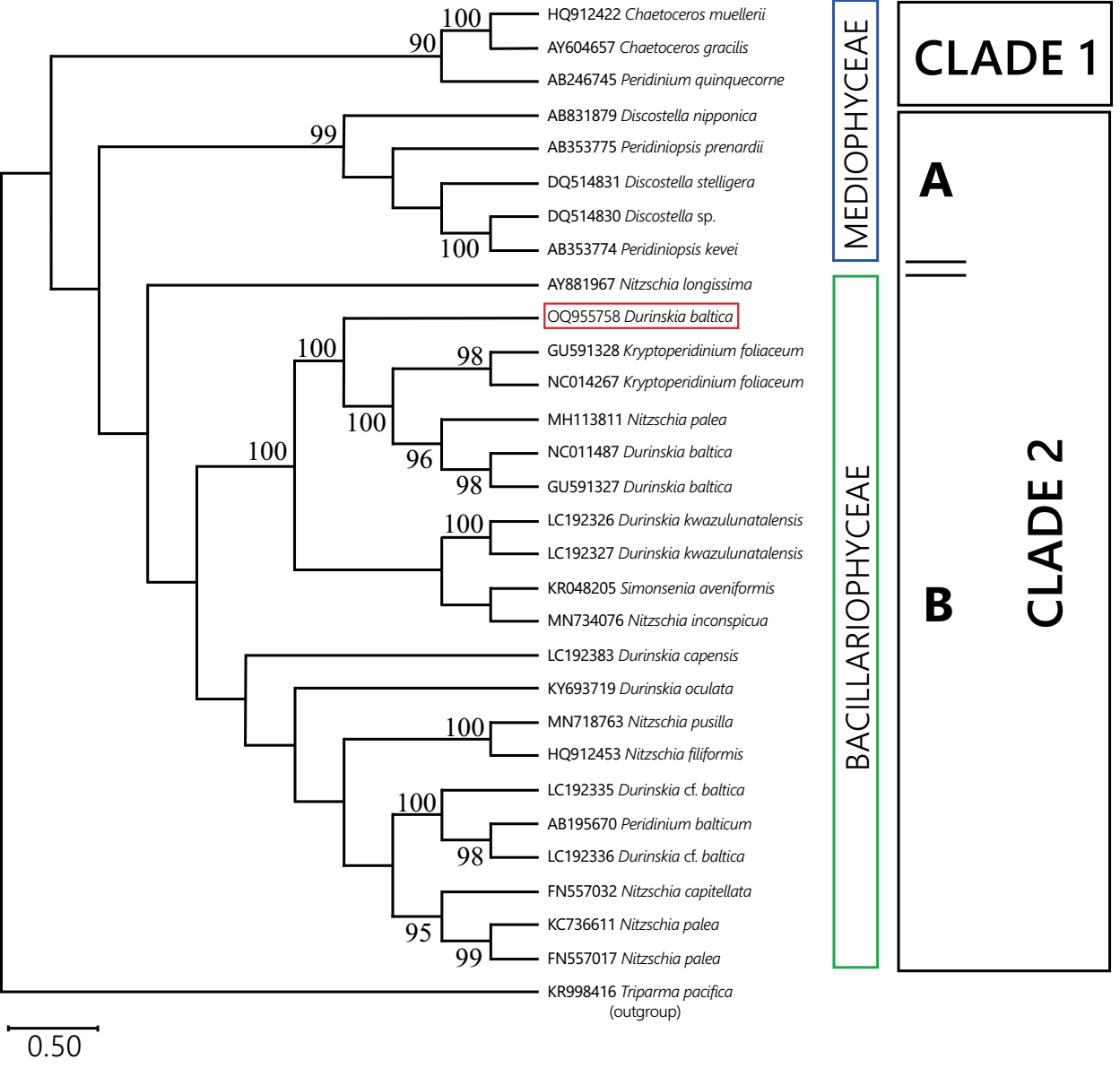
Figure 6 Bayesian phylogeny of rbcL sequences of dinotome endosymbionts and diatoms. Endosymbionts sequences are named after the host to which they belong. Numbers next to nodes correspond to Bayesian posterior probability values. For each clade, the class-level membership of the current diatom sequences is indicated. Clade 1 assembled diatom and endosymbiont sequences from marine habitats. Clade 2 assembled sequences from freshwater (Mediophyceae) and marine or freshwater (Bacillariophyceae) diatoms and endosymbionts. Endosymbiont of Durinskia baltica from Mexico framed in red.
Discussion
Morphology. The cell shape in dinotoms of the genus Durinskia from continental environments varies, ranging from globular to slightly dorsoventral. In the literature, this feature is considered necessary for describing the species (Hansen & Flaim 2007, Cavalcante et al. 2017, Lira et al. 2017, Kretschmann et al. 2018, Moestrup & Calado 2018, Lira & Tavera 2019). In our material, there are specimens with spherical cells and ellipsoidal cells, both slightly dorsoventrally flattened.
In populations of Durinskia isolated from freshwater environments, including those in this study, there are smooth plates with few delicate and minute pores, scattered without order, and some aligned parallel to the plate margin (Hansen & Flaim 2007, Lira et al. 2017, Kretschmann et al. 2018, Lira & Tavera 2019). In the genus systematics, size, shape, and dorsoventral flattening are variable (Moestrup & Calado 2018). Suppose the presence of delicate pores were the diacritic character. In that case, there is no clear morphological difference between D. baltica from Mexico and D. oculata from Prague (Moldava River, the type locality of its basionym Glenodinium oculatum Stein) (Kretschmann et al. 2018). Regarding the genetic basis, the similarity of their sequences in the 18S rRNA phylogeny (Figure 5 Table S2, Supplementary material) would certainly support their synonymy. Their endosymbionts, on the other hand, do not appear to be so clearly related (Figure 6, Table S3), so if the endosymbiosis occurred with different species of diatoms and perhaps in different regions and environments, there would not be a solid basis for using a single name at the moment. As we explained before, we decided to retain the name D. baltica for the population of Mexico.
Genetic Analysis. Analysis of the host sequences of the Durinskia population from Mexico generated an 18S rRNA phylogeny for dinotoms that is consistent with those presented by several authors, including the character of monophyly for Kryptoperidiniaceae (Horiguchi & Takano 2006, Moestrup & Daugbjerg 2007, Yamada et al. 2017, Kretschmann et al. 2018). The morphology of our population supports monophyly by showing that it possesses the apomorphic characters that distinguish it, i.e., the endosymbiont core, the hooked stigma, and the presence of only two intercalary plates (Pienaar et al. 2007, Takano et al. 2008, Lira et al. 2017, Kretschmann et al. 2018). The possible time of origin of dinotoms through molecular calibration of sequences has yet to be fully explored in Kryptoperidiniaceae. There are time estimates for Gymnodiniaceae and Peridiniales, made to date marine to freshwater transitions. Since the sample from Mexico corresponds to an epicontinental environment, the molecular dating of the phylogenetic trees for the sequences was consistent with the information known so far (Žerdoner-Čalasan et al. 2019).
The phylogeny calibrated in our study also supports the postulate that, within each family, brackish and freshwater species had a more recent origin than marine species (Figure 5). The location of the ancestors of marine, brackish, and freshwater dinophytes that calibrated phylogeny suggests near the Ordovician-Silurian boundary (443.8 mybp) supports the transitions that have been proposed from secondary to tertiary endosymbiosis as having an essential role in the history of dinotomes and other dinophytes (Pienaar et al. 2007, Hehenberger et al. 2014, Ishida & Green 2022). As far as is known, dinotomes with a tertiary endosymbiont evolved from an ancestor with secondary endosymbiosis containing peridinin. The only remnant of the ancestral secondary endosymbiosis is preserved in the characteristic stigma they possess. Fucoxanthin, as in diatoms, is the carotenoid produced in the present-day chloroplasts of dinotoms (Pienaar et al. 2007, Hehenberger et al. 2014, Bai et al. 2022, Ishida & Green 2022).
Other studies on dinophyte antiquity posit the marine-freshwater transition of several species (Fensome et al. 1996, Žerdoner-Čalasan et al. 2019). Also, consistently, our analysis presented significant diversification for freshwater species after the Cretaceous-Paleogene boundary (66 mybp) indicated as the K-Pg boundary (Figure 5). Branches representing freshwater sequences formed a strongly supported sister relationship with branches from marine sequences, which agrees with previous literature (Gottschling et al. 2017, Žerdoner-Čalasan et al. 2019). As mentioned by Kretschmann et al. (2018), the isolated arrangement of the freshwater sequence groups in the phylogenies suggests that there is little relationship between them; at the same time, their closest relatives within each family are marine or brackish origin (Figure 5).
The sample from Mexico belongs to a freshwater environment, and the freshwater dinotom species in Kryptoperidinaceae, Peridiniaceae, and Peridiniopsidaceae have endosymbionts whose sequences are similar to diatoms such as Discostella, Thalassiosira, and Cyclotella, or Nitzschia-like, all from freshwater (Figure 6).
The genetic distance between Durinskia sequences from saline and freshwater environments was robust (BPP = 1.0), and they could have diverged by more than 250 mybp (Figure 5). The most recent origin for freshwater dinophytes of this genus was indicated by the sequences of D. oculata from Prague (type locality, sequence KY693722) and D. baltica from Xochimilco (sequence OQ867280), both 20 mybp. They showed a close genetic relationship and shared a common ancestor with other freshwater sequences, such as sequence GU999528 of D. baltica from China (Figure 5 Table S2, Supplementary material).
The results suggest that, by contrast, their endosymbionts have accumulated a different rate of change, implying a different degree of differentiation (Figure 6, Table S3). Other studies have mentioned this observation (Zhang et al. 2011, Kretschmann et al. 2018), but further research is needed to establish how many alleles their genomes share and how much synteny exists, as in diatoms it is highly variable (Crowell et al. 2019). In this study, it was clear from the BPP values that the Bayesian phylogeny result was not informative enough to support a molecular calibration for endosymbionts (Figure 6). Also, exploring the rbcL sector of extant diatoms associated with dinotome endosymbionts revealed that genetic relationships among Nitzschia species are unclear (Figure S1), which may have contributed to weakening the endosymbiont analysis (Sorhannus 2007).
As pointed out by Žerdoner-Čalasan et al. (2018), "endosymbionts of Kryptoperidiniaceae found their closest relatives in free-living diatoms and not in other algal hosts." Sequences from Durinskia baltica from Mexico and D. oculata from Europe showed the same (Figures 5, 6, Tables S2, S3). Their endosymbionts do not share a close relationship (Zhang et al. 2011, Kretschmann et al. 2018). A comparable case was indicative in the marine dinotoms D. capensis and D. kwazulunatalensis, which have similar morphology and phylogenetic relationship and harbor distantly related endosymbionts. However, we do not regard it with equal importance because, in these two species, the endosymbionts have a different degree of incorporation with the host; in D. capensis, it is a temporary endosymbiont incorporated through kleptoplastidy, and in D. kwazulunatalensis the host incorporation is incomplete because it possesses several endosymbiont nuclei, not just one (Yamada et al. 2017, 2019). According to Moestrup & Daugbjerg (2007), those species with incomplete endosymbionts might be uninformative for phylogenies.
Our results support the proposed diversity of endosymbionts occurring in dinotoms, which belong to at least five diatom genera, according to Yamada et al. (2017). Although some host-endosymbiont genetic integration has not been proved in Durinskia, differences at the specific level of putative endosymbionts (Nitzschia type) may be considered in the systematics of this genus concerning the cytosolic activity of hosts that is not yet sufficiently studied (Hehenberger et al. 2016, Janouškovec et al. 2016).
Several authors point out that in most dinotoms, especially marine dinotoms, the tertiary endosymbiont is of Nitzschia type and suggest that the replacement of the endosymbiont (and hence the current diversity) linked to the evolutionary history of dinophytes (Horiguchi & Takano 2006, Takano et al. 2008, Imanian et al. 2010, Hehenberger et al. 2016, Yamada et al. 2017). In the case of D. baltica from Mexico, endosymbiosis may have occurred in a brackish continental environment and subsequently transited to freshwater because Xochimilco was part of a large brackish lacustrine area (Figure 1). The current freshwater condition appears to have begun about 8,000 years ago (Ortega-Guerrero et al. 2018). In our calibrated phylogeny (Figure 5), the origin of the Mexican dinotom places 20 mybp, and the geological origin of the Valley of Mexico basin to which Xochimilco belongs is 26 mybp (Arce et al. 2019). It is not precludable that some replacement in the endosymbiont diatom type would have occurred in the evolutionary history of D. baltica from Xochimilco. According to Yamada et al. (2017), there are records of Nitzschia species replacements in several dinotoms.
One observation that stands out in that evolutionary history is that currently, Durinskia baltica from Mexico corresponds with a dinotom that controls the karyokinesis of its guest and inherits it to daughter cells, i.e., it is not the kleptoplastid or incomplete model proposed in the literature (Yamada et al. 2017, Kretschmann et al. 2018, Yamada et al. 2019). The character of the endosymbiont as definitive is strongly supported by previous studies of the Mexican population in culture, with light microscopy, epifluorescence, and transmission electron microscopy observations, which confirmed that a single membrane surrounds the single nucleus, chloroplasts, and ribosomes of the endosymbiont, permanently separating it from the host's cytoplasm; this structure is not modified during the life cycle and inherits by daughter cells (Lira et al. 2017 [Figure 3], Lira & Tavera 2019).
We conclude that the calibrated phylogeny reflected the proposed general times of origin for dinotomes and allowed us to temporally place the proposed evolution of the group about the acquisition of tertiary endosymbionts. For example, Durinskia, Peridinium, Peridiniopsis, and Kryptoperidinium possess diatom-like endosymbionts whose evolutionary line separates them from those with a haptophyte-like endosymbiont. Therefore, it was crucial to place the Mexican species in the dating context that has been proposed in the literature for marine and freshwater dinotomes. Notably, the focus of this study highlighted that the evolutionary transition of marine dinophytes to brackish or freshwater proposed in the literature is geologically contextualized in the case of the Xochimilco basin, suggesting the occurrence of colonization of D. baltica in a brackish environment and its posterior adaptation to freshwater was consequent to the environmental change of Xochimilco Lake for salinity.
Likewise, the calibrated phylogeny indicates that freshwater Durinskia populations share a common ancestor and could have diverged before colonizing freshwater around the K-Pg boundary as proposed in the literature; this also applies to other freshwater dinotomes of Peridinium and Peridiniopsis (Figure 5). The differences between the endosymbiont diatom types are a product of each particular environment colonized by the dinotoms and is an exciting line of research going forward.
Supplementary material
Supplemental data for this article can be accessed here: https://doi.org/10.17129/botsci.3259











 nueva página del texto (beta)
nueva página del texto (beta)



