Many organisms, including nematodes, gastropods, insects and vertebrates, use plants as a food source. Plant tissues are an important source of carbohydrates, lipids and proteins to animals, but plants also produce non-nutritive substances known as secondary metabolites. Despite the wide distribution of phytochemicals across plant taxa, they were initially considered waste products from primary metabolic pathways, since they lack an “apparent” metabolic function (Pichersky & Lewinshon 2011). However, this idea was abandoned after the work of the entomologists Dethier (1941, 1954) and Fraenkel (1959) (see also Harborne 2014) demonstrated that secondary metabolites are related to the acceptance of plants as food by herbivores, acting either as attractants or repellents. Since then, the ecological role of secondary metabolites as mediators of biotic interactions (e.g., herbivory, pollination) has been extensively documented (Ode 2006), and the interaction between plants and herbivores became the scaffolding on which major hypotheses of biological diversification and evolution are based (Stamp 2003, Agrawal 2006). In particular, the “escape and radiation” hypothesis placed the plant-herbivore interaction, or more precisely, the reciprocal coevolutionary responses of plants and herbivores, as the ultimate drivers of speciation and diversification (Ehrlich & Raven 1964, Janz 2011).
Evolutionary history of plant-herbivore interaction. Origins of interaction.- Plant-herbivore interaction is one of the largest and most complex interactions among organisms. From microorganism to vertebrates, all kind of animals feed exclusively on plant tissues, and the sum of green plants (~25-30 %) and phytophagous insects (26 %) accounts for nearly half of the known species on the planet (Strong et al. 1984, Llorente-Bousquets & Ocegueda 2008). The diversification patterns of plants and herbivores found their origins during the Paleozoic, 300-400 million years ago (Myr), when the first vascular plants and arthropods emerged (Scott et al. 1992, Price 2002). Fossil records of leaf damage, coprolite dispersion, specialized mouthparts and intestinal contents of orthopterans, evince the antagonistic interaction between plants and arthropods (Labandeira 1998, 2007, Iannuzzi & Labandeira 2008). Besides documenting structures associated with these interactions (e.g., Labandeira 1997), the fossil record allows the sequence of emergence events of plant and animal taxa to be followed and supports the first well-accepted major (macro) evolutionary patterns. Records suggest that the evolution of vascular plants, dating back to the Devonian, about 350-400 Myr (but see Box 1), preceded the evolution of arthropods during the Carboniferous, 300 Myr. During the late Paleozoic and early Mesozoic 250 Myr, important groups of phytophagous insects such as Coleoptera and Lepidoptera appeared. Then, around 100 million years later, the angiosperms emerged and widely diversified around the world (Niklas 1983). Hence, these major patterns have contributed to the idea that diversification of plants and herbivores (particularly insects) is, in fact, a result of the biotic interactions between plants and animals (Strong et al. 1984, Nylin & Janz 1999). For instance, it has been suggested that both the “explosive diversification” and dominance of the angiosperms during the Cretaceous were detonated by, or at least associated with, animal-mediated pollination (Faigri & Van der Pijl 1979). In addition, it is argued that zoophilic pollination may have originated from a pollen-feeding behavior of animals on early angiosperms, probably evolving during the Jurassic, from coleoptera feeding on spores in free spore plants (e.g., Cycadeoideales, Scott et al. 1992).
Box 1. The molecular clock and the earlier colonization of land
By comparing mutation rates of biomolecules of different lineages, either nucleotide sequences for DNA or amino acid sequences for proteins, it has been possible to deduce the time at which two or more life forms diverged. The molecular clock hypothesis asserts that the rate of evolutionary change of any protein is approximately constant over time, thus over differ lineages (Kumar 2005). This prediction however, has run into certain challenges at different time scales (e.g., saturation; accumulation of molecular evolutionary changes through time that flattens out linearity, Marshall et al. 2015). However, even considering its limitations, the molecular clock has significantly contributed to dating phylogenetic events, including those that are not fossil-based. A relatively recent study based on molecular clock analysis suggests that early colonization of land is older than current fossil-based estimates (Heckman et al. 2001). The first fossil land plants and fungi appeared 480-460 Myr, whereas analysis of protein sequences of alga and fungi sets the emergence date 1,000 Myr. It has been suggested that colonization of the earth by eukaryotes was probably facilitated by the symbiotic relationship between a photosynthetic organism and a fungus. And, based on molecular clock estimates, an earlier colonization of land, about 600 Myr. In the same fashion, molecular clock estimates, in addition to evidence of morphological diversity of flower, seeds and pollen from the mid Cretaceous and the presence of derived lineages from increasingly older geological deposits, suggest that the timing of early angiosperm cladogenesis is older than fossil-based estimates have indicated (Wikström et al. 2001). Nevertheless, colonization time hypotheses of major lineages based on divergence of DNA data remain speculative.
Early functions of secondary metabolites. Paleontological analyses have clearly shown the association of early vascular plants and arthropods, but do not necessarily account for the chemical substances (e.g., secondary metabolites) involved in the interaction (e.g., Scott et al. 1992). In this context, hypotheses attempting to explain the evolution of the chemical response of plants to defoliation as a defensive strategy based on morphological analysis of fossils (e.g., the pattern of leaf feeding by arthropods), remain speculative (see Cooper-Driver 1978, Scott et al. 1992). Perhaps the most compelling evidence of early (defensive) function of secondary metabolites are derived from biochemical analyses of ancient and/or living angiosperms (e.g., Weng & Chapple 2010, Paul & Dutta 2017) and from the use of chemistry and biochemistry in comparative morphological analyses of plant systematics.
Particular properties and functions distinguish secondary metabolites from those derived from the primary metabolism (Box 2). Since the first secondary metabolites were produced by aquatic organisms, it is suggested that their early function might have been related to excretion mechanisms derived from the incomplete cycling of primary compounds soluble in water (McClintock & Baker 2001). Then, as organisms and their interactions became more complex, secondary metabolites acquired different functions (e.g., conspecific chemical communication; Lovett et al. 1989, Baluška & Ninkovic 2010). It is possible that the defensive function of secondary metabolites of photosynthetic eukaryotes may have been triggered by one evolutionary milestone: the transition from aquatic (presumably non-vascular) plants to vascular terrestrial plants (tracheophytes) during the Silurian (but see Heath 1987, 1991). Geochemical analysis shows that after the appearance of the first terrestrial vascular plants, a very rapid biochemical diversification occurred, involving in some cases fundamental biogenetic changes (Niklas 1983).
Box 2. Secondary metabolite biosynthesis
Secondary metabolites are structurally and functionally diverse organic molecules not involved in primary metabolic functions of living organisms. Most secondary metabolites affect more than one important biological process, usually related to survival and interaction with the environment (e.g., flavonoids are involved UV light protection, and defense against plants’ natural enemies; Treutter 2006, War et al. 2012, Adeboye et al. 2014). Unlike primary metabolites, secondary metabolites have a restricted taxonomic distribution (particular compounds often occur only in single genus or species), and the machinery and pathways needed to produce different classes of compounds are highly conserved (i.e., closely related species share more traits or ecological associations than do distantly related species; Wink & Mohamed 2003, Agrawal 2006).
Secondary and primary metabolism are intersected by diverse products of primary biosynthetic pathways, which entails the “starter units” of secondary biosynthesis. The main starter units are Acetil CoA and pyruvic acid. Once they are combined with “extender units” such as malonic CoA, mevalonic acid, or erythrose 4-phosphate, among others, the synthesis of diverse secondary pathways is triggered. Prime biosynthetic routes include: (1) the shikimate or shikimic acid pathway, which is the biosynthetic route to the aromatic amino acids phenylalanine, tyrosine, and tryptophan. This pathway has been found only in microorganisms and plants, and is a bit more widespread than nitrogen fixation or photosynthesis but less ubiquitous than, for example, nitrogen assimilation (Herrmann 1995). (2) The mevalonate or the isoprenoid pathway is the route by which terpenes and steroids are synthetized. This pathway is present in eukaryotes, archaea and some bacteria. By the binding of five-carbon isoprene units, this route is responsible for over 30,000 biomolecules, including cholesterol, vitamin K, and all steroid hormones. (3) The acetate- malonate pathway is the route of fatty acids and polyketides. This pathway is present in bacteria, fungi and plants. Through this route a great diversity of secondary metabolites are synthetized, including aromatic and aliphatic compounds, prostaglandins and flavonoids, among others (Dewick 2002).
Lignification is a key adaptation for life in a terrestrial environment, and entails one of the most striking biosynthetic changes that came with plant vascularization. Lignin is a complex cross-linked phenolic biopolymer made up of several different monomers such as coniferyl alcohol and sinapyl alcohol. Besides its well-known structural properties, lignin regulates the hydration of the hydrophilic molecules in the cell wall and provides excellent protection against ultraviolet light. In general, phenolic acids, derived from the aromatic amino acids of algae, function as absorption agents of ultraviolet light. For this reason, the evolution of lignin is attributed to the increasing exposure to UV light of early tracheophytes (Lowry et al. 1980, Weng & Chapple 2010). Hence, it is proposed that the chemical evolution of lignin was possible through the reductive deamination of some aromatic amino acids such as tyrosine, present in algae, since the p-coumaril residue produced by the deamination of tyrosine is a more effective compound as a UV light filter than tyrosine itself. In addition to UV light protection, several phenols react synergistically with hormones and are able to inhibit ATP synthesis, disengaging respiration or interfering with ion absorption, functioning as a formidable barrier to pathogens (Nicholson & Hammerschmidt 1992, Adeboye et al. 2014). Phenolic derivatives include tannins and flavonoids, two significant classes of defensive secondary metabolites of plants. Tannins are usually found in high concentrations in plants, sometimes constituting more than 60 % of the dry weight of a tissue. Along with specific proteins, tannins built complexes that are very difficult to digest and that reduce the amount of nitrogen that can be harnessed by proteins. Thus, tannins inhibit the digestive enzymes of animals and reduce the ability of pathogens to hydrolyze proteins (Barbehenn & Constabel 2011). Flavonoids are some of the few secondary metabolites synthesized by all vascular plants (and some algae). Flavonoids are responsible for many plant attributes, such as pigments and flavor, and like most phenolic compounds, prevent pathogen invasion and affect the activity of digestive enzymes of animals (Treutter 2006, War et al. 2012). Paleobiochemical analyses of Devonian and low Carboniferous plants, and of angiosperms from the Cenozoic, demonstrate an impressive increase in biochemical diversity, which includes lignin derivatives, terpenoids, tannins and flavonoids, among others (Cronquist 1977). These findings suggest that although metabolites’ evolutionary origin may be associated with abiotic factors such as UV light, their diversification might be related to the emergence of the angiosperms and the first arthropods (and the interaction between them). This means that ca.30 Myr after the emergence of primitive lignin (during the late Silurian and early Devonian), most plant secondary metabolites were probably functioning as mediators of early plant-herbivore associations, similar to current biological associations.
Ecological role of secondary metabolites. Despite immense diversification in plant-herbivore interactions over the past 450 million years, the preference of some herbivores for particular host plants lingers. For instance, Danaus plexippus (Lepidoptera: Nymphalidae) larvae preferentially feed on Asclepias spp., which is known to have cardenolides (Malcolm & Brower 1989, Agrawal & Konno 2009). It is precisely the constancy of some interactions apparently mediated by secondary metabolites which has motivated a large number of studies trying to elucidate the ecological role of chemical compounds. Many studies have in fact shown that current associations between plants and herbivores are often related to the presence of a particular kind of phytochemical (e.g., Cornell & Hawkins 2003, Theis & Lerdau 2003). These compounds act as repellents or attractants of diverse herbivore groups. For instance, larvae of Spodoptera eridania (Lepidoptera: Noctuidae) prefer to feed on plants containing cyanogenic glycosides, and appear to grow better when cyanide is present in their diet (Brattsen et al. 1983). Nevertheless, most work is focused on the defensive role of secondary metabolites, describing a wide range of negative effects on plant consumers.
Three basic biosynthetic pathways are responsible for most of the secondary compounds produced by plants (Box 2). It is estimated that plants are able to produce around 100,000 different chemical compounds, including terpenes, alkaloids, steroids, phenols, and glycosides, among others (Table 1), some of which may not be involved in a particular interaction with a plant enemy (Dixon 2001). Two criteria must therefore be met in order to consider a plant character defensive (Karban & Baldwin 1997). First, the attribute must affect the extent to which a plant is attacked (e.g., reduce leaf damage), and second, it must have a positive effect on some component of plant fitness (i.e., fecundity, mating success or survival). Within species, many studies using quantitative genetics or measuring natural selection have shown the defensive role of secondary metabolites. For instance, Castillo et al. (2014) addressed the pattern of phenotypic selection on the secondary compounds atropine and scopolamine in a natural population of Datura stramonium, a solanaceous weed known to have tropane-derived alkaloids. They showed a positive effect of tropane alkaloids on plants’ fecundity. In the same species, an experimental analysis of the association of plant compounds with herbivore damage showed a negative correlation between a precursor of scopolamine (hyoscyamine) and the leaf damage inflicted by generalist herbivores (Shonle & Bergelson 2000).
Table 1 Biosynthetic pathway and taxonomic distribution of major secondary metabolites
| Chemical compound | Biosynthetic pathway (in plants) | Taxonomic distribution | Reference |
|---|---|---|---|
| Alkaloids | Shikimic acid | ~20 % of angiosperms (commonly found in Leguminosae, Liliaceae, Solanaceae, Papaveraceae, Apocynaceae, Amaryllidaceae y Rununculaceae), microorganism, cyanobacters, algae, marine invertebrates (bryozoans, porifera, molluscs, coelenterates and tunicates), amphibians and arthropods (insects) | Culvenor 1973 Harborne 2014 Waterman 1993 Daly et al. 2005 Fattorusso &Taglialatela-Scafati 2008 Grindberg et al. 2008 |
| Cardiac glycosides | Acetate-mevalonate | In 12 taxonomic families. Mainly in Apocynaceae / Asclepidaceae and Cruciferae | Gershenzov & Croteau 1991 Malcolm 1991 |
| Cyanogenic glycosides | Shikimic acid | In most vascular plants; Gymnosperms, Angiosperms (monoctodilenoeas and dicotyledons) | Seigler 1991 Bennett & Wallsgrove 1994 |
| Glucosinolates | Shikimic acid | Widely distributed in Brassicales/Capparales (larger presence in Brassicaceae, Capparidaceae and Tropaolaceae | Louda & Mole 1991 Bennett & Wallsgrove 1994 Mitöfer & Boland 2012 |
| Latex | Different biosynthetic pathways | ~ 10 % of Angiosperms, and Mushrooms | Mitöfer & Boland 2012 |
| Phenols | Mixed biosynthetic pathway | Widely distributed in vascular plants. Tannins and lignin are not found in animals | Hagerman & Butler 1991 |
| Phytohormones | 1. Fatty acids route (linolenic acid) 2. Shikimic acid | Widely distributed in angiosperms and gymnosperms. *It has been discussed whether they should be considered primary metabolites | Bennett & Wallsgrove 1994 |
| Non-Protein amino acids | Bounding of a carboxyl group (-COOH) and an amino group (-NH2) | Mainly distributed in Leguminosae and Papilionaceae | Rosenthal 1991 |
| Steroids and terpenes | Acetate-mevalonate | Widely distributed in angiosperms Steroids commonly found in Caryophyllaceae, Leguminosaceae, Sapindaceae, Liliaceae, Dioscoreaceae | Harborne 2014 Gershenzov & Croteau 1991 |
Besides analyzing the effects of phytochemicals on different animal species, many studies have also contributed to detail their mode of action on several biological systems, such as cell metabolism and enzymatic activity. Analysis of the interaction of enzymatic complexes with compounds derived from amino acids has shown, for instance, that alkaloids affect plant consumers by inhibiting enzyme activity and competing for muscarinic acetylcholine receptors (Roddick 1991, Grindberg et al. 2008). In addition, the study of structure and biosynthesis of secondary metabolites and the analysis of their distribution across plant taxa have contributed to examine the specificity of plant-herbivore interactions and to developing hypotheses focused on explaining the evolutionary process that has led to diversification of plants and animals.
The biosynthesis of flavonoids is perhaps one of the best understood biosynthetic routes and is considered one of the major experimental models for the study of enzymatic machinery. Recent work on Arabidopsis sp. for instance, has provided new information on the channeling of intermediaries and the assembly of multienzymatic complexes (Winkel-Shirley 2001, Bowerman et al. 2012). Also, evolutionary studies of the phenilpropanoids synthesis of Ipomoea purpurea have suggested that enzymes involved in the biosynthesis of flavonoids were derived from enzymes of the primary metabolism, and that gene duplication has allowed the adaptation of these enzymes to specific functions (Rausher et al. 1999, Durbin et al. 2000). This evidence links the evolutionary history of chemical compounds with their current functions, thus forming a “bridge” between intra- and interspecific approaches. Through the analysis of nucleotide sequences, including structural genes, regulatory sequences, and noncoding DNA segments within chromosomes, the common origin (and function) of genes across species can be assessed, at the same time as the specific genetic attributes of a single taxa (Chen et al. 2016).
Overall, empirical evidence at the microevolutionary scale derived from ecological, taxonomic, biochemical and genomic studies, provides strong support for the role of secondary metabolites as mediators of plant-herbivore interactions, particularly as a defense against all types of herbivores (Table 2).
Table 2 Function of major secondary metabolites in plants.
| Chemical class | Example | Plant taxa | Effect | Mode of action | Target taxa | Reference |
| Alkaloids | Solanine | Solanum demissum (Solanaceae) “potato” | 1. Toxic 2. Deterrent | Compete for muscarinic acetylcholine receptors | * Empoasca fabae * (Hemiptera: Cicadellidae) “green grass leafhopper” * Leptinotarsa decemlineata (Coleoptera: Chrysomelidae “potato beetle” | Bennett & Wallsgrove 1994 Roddick 1991 |
| Hyoscyamine | Datura stramonium (Solanaceae) “jimsonweed” | 1. Deterrent | n. d. | * Epitrix sp. (Coleoptera: Chrysomelidae) “potato flea” | Shonle & Bergelson 2000 | |
| Cardiac glycosides (Steroids) | Cardenolides | Asclepias sp. (Apocynaseae) “milkweed” | 1. Toxic and deterrent (bitter taste) 2. Attractor | Inhibit Na+ pump in cardiac, vascular and nervous cells of animals | * Arthropods (particularly insects). Asclepias- Danaus sp – predatory birds. When larvae of Danaus sp feed, acquires the cardenolides that are used as a defense against predatory birds * Danaus plexippus (Lepidoptra: Nymphalidae)” monarch butterfly ” | Malcolm 1991 Agrawal 2005 Brower et al. 1975 |
| Cyanogenic glycosides | Cyanide (Released by cyanogenesis) | Manihot esculenta (Euphorbiaceae) “cassava” | 1. Toxic (but see Brattsen et al. 1983) | Affects cellular respiration by inhibiting oxygen from binding with the enzyme cytochrome-c-oxidase) | * Cyrtomenus bergi (Hemiptera: Cydnidae)”smallpox bug” | Bennett & Wallsgrove 1994 Mitöfer & Boland 2012 |
| Glucosinolates | Total content by species | Brassicaceae | 1. Toxic and deterrent 2. Attractor and phage stimulant | n. d. | * Lipaphis erysimi (Hemiptera:Aphididae) “turnip aphid” * Deroceras reticulatum (Gastropoda: Agriolimacidae)” gray fieldslug ” * Psylliodes chrysocephala (Coleoptera: Chrysomelidae) “cabbage-stem flea beetle ” | Bennett & Wallsgrove 1994 Rhoades & Cates 1976 |
| Latex | Total content by species | Hoodia gordonii (Apocynaceae) “hoodia” | 1. Toxic and deterrent 2. Antinutritive 3. Sticky | n.d. | * Trichoplusia ni (Lepidoptera: Noctunidae) “cabbage looper” * Arthropods mainly insects | Chow et al. 2005 Agrawal & Konno 2009 |
| Phenols | Tannins | Quercus robur (Fagaceae) “English oak” | 1. Antifeedant 2. Toxic | Alters digestive enzyme activity | * Operophtera brumata (Lepidoptera: Geometridae) “winter moth” | Feeny 1970 |
| Flavonoids [flavonol 3-(2”-xiloxygalactosido)] | Crucifers | 1. Attractor and phage stimulant | n. d. | * Phyllotreta armoraciae (Coleóptera: Chrysomelidae) ”Horseradish flea beetle” | Nielsen et al. 1979 | |
| Phytohormones | Salicylic acid | Nicotiana sp (Solanaceae) “tobacco” Brassica napus (Brassicaceae) “rapeseed” | 1. Resistance induction to pathogens 2. Synthesis of defense induction (e.g. glucosinolates) | Induces proteins (PR) and enzymes such as 2-hydroxylase benzoic acid. | * Tobamovirus (TMV) “tobacco mosaic virus” Several arthropods, particularly insects. | Bennett & Wallsgrove 1994 |
| Non-protein amino acids | Canavanine | Legumes | 1. Toxic 2. Deterrent | Is incorporated into protein synthesis, having substantial deleterious effects. | * Caryedes brasiliensis* (Coleoptera: Bruchidae) “low-necked beetle“ * Sternechus tuberculatus* (Coleoptera: Curculionidae) * They have the ability to discriminate between arginine and canavanine, reducing the incorporation of canavanine into the system | Rosenthal et al. 1976 Rosenthal 1986 |
| Steroids and Terpenes | Gossypol | Gossypium sp (Malvaceae) “cotton” | 1. Toxic 2. Antifeedant | Inhibits enzymatic activity | Arthropods: * Aphis gossypii (Hemiptera:Aphididae) “cotton aphid” * Lygus hesperus (Hemíptera: Miridae) “western plant bug” * Estigmene acrea (Lepidóptera: Actiidae) “the saltmarsh caterpillar” Fungus * Verticillium dahliae | Bottger et al. 1964 Bennett & Wallsgrove 1994 |
n.d., non-determined
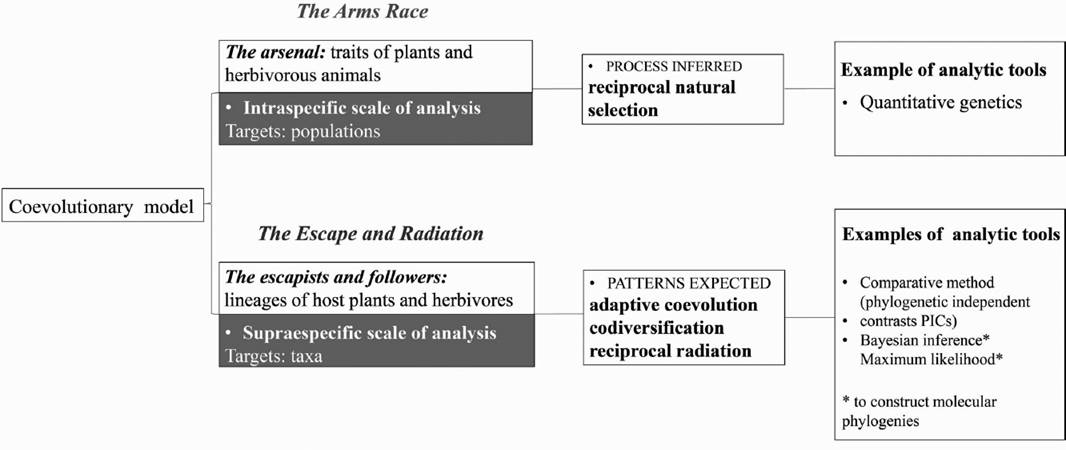
The coevolutionary model of Escape and Radiation and the Arms Race. Identifying common evolutionary patterns among host plants and their hosts has been one of the main goals of most early studies of plant-herbivore interactions. Based on ecological data from plant-insect associations, Ehrlich & Raven (1964) developed the conceptual framework for the study of plant-herbivore interaction. The authors documented a conserved phylogenetic pattern of the interaction between host plants and butterflies, mediated by secondary metabolites characteristic of the host plants. These phylogenetic constraints were placed in an explicit theoretical context, which was later known as the “escape and radiation” hypothesis. They proposed a coevolutionary model based on (1) the evolution of novel defensive traits in plants, in response to selective pressure imposed by their consumers, and (2) the evolution of counter-defenses in the consumers. This “arms race” (Dawkins & Krebs 1979) model portrays one of the ways in which coevolution can occur (Thompson 1989, 1994), and describes how reciprocal selection can lead to plants escaping from their herbivores through mutations (novel defenses), resulting in adaptive radiation. Consequently, herbivores able to surpass the novel defenses could thus radiate on diversified host plants. This “escape and radiation” hypothesis predicts corresponding speciation patterns.
Ehrlich and Raven’s metaphors of coevolution encompass both the mechanism and outcomes of evolution, and they function at different hierarchical scales (Figure 1). Metaphor 1: the “arms race” represents the explanatory process of patterns that emerged either at ecological or phylogenetic scales. These predictions are usually tested within populations by means of trait-based analyses. Metaphor 2: the “escape and radiation” represents the outcome of the “arms race”, i.e., the phylogenetic patterns. Thus, this prediction is only tested across phylogenies, and frequently uses taxon-based analyses. Host shifts related to plant chemical attributes and phylogenetic correspondence of plant and herbivore taxa documented for certain lineages, are perhaps the best empirical evidence supporting this part of the model. However, it is unclear how counter–defense coevolution resulted in adaptive radiations. Some of this debate is further discussed below.
The “arms race”: Defense vs. counter-defense. Defensive and counter-defensive strategies have usually been assessed in the context of classical defense theory (Stamp 2003, Agrawal 2006), derived from the “arms race” model. From this perspective diverse plant attributes are associated with defense strategies, including: (1) seasonal distribution of nutrients and storage in diverse parts of the plant (Hunter et al. 1992), (2) physical traits and chemical compounds that avoid or reduce leaf damage by herbivores (i.e., resistance; Painter 1958, Strauss & Agrawal 1999), and (3) the ability to buffer the impact of leaf damage on fitness (i.e., tolerance, van der Meijden et al. 1988). In addition, plants have a set of structural defenses such as the cuticle, cell wall and stomata, which represent a physical barrier to microorganisms and pathogens (Melotto et al. 2006, 2008). Physical resistance includes waxes on the leaf surface, thorns, leaf trichomes, hardness and leaf folding. Chemical resistance includes a diverse array of compounds such as tannins, alkaloids, phenols, and glycosides, among others.
Resistance traits can be expressed constitutively or induced (i.e., increasing after the plant is attacked, Karban & Baldwin 1997) by pathogens, nematodes, invertebrates or phytophagous vertebrates (Walters 2011). The “machinery” required for the deployment of plants’ induced defenses can be very complex. For instance, recognition paths of pathogens or nematodes involve a series of signals that are activated when the cuticle and cell wall break. When a plant is attacked by pathogens, recognition receptors located on the surface of cells are activated. When the PAMP (perception of pathogen-molecular patterns) mechanism is activated by recognizing pathogen-associated molecules such as chitin, Gram-negative bacterial lipopolysaccharides, and peptides derived from bacterial flagella (Zipfel 2008), the immune system is triggered, and is the first level of immunity in response to pathogens. Other inducible defenses in response to pathogen attack include the formation of cork layers, lignification and production of chemical compounds such as phytoalexins (Ahuja et al. 2012, War et al. 2012). Besides pathogens and nematodes, chemical defenses of plants are usually induced by herbivorous insect attack. Plants of the genus Bursera produce a set of monoterpenes that are distributed in a network of channels in the stems and leaves. Leaf damage results in a rapid release of fluids toxic to herbivores at the site of damage (Becerra & Venable 1990, Becerra et al. 2001). Other substances released after the attack of herbivores such as some resins in latex, besides being toxic, may reduce digestibility (Agrawal et al. 2008) or inhibit growth (Konno et al. 2004). However, since plants simultaneously interact with a diverse community of consumers, usually a single plant taxon has a diverse array of defensive characters, including several chemical compounds. In addition, a single compound may have different effects on different consumers (e.g., repellent, attractant, see Izhaki 2002). Table 2 shows the effect and mode of action of the main classes of plant secondary metabolites on diverse herbivores.
It has been suggested that depending on the predictability of their distribution (i.e., “appearance”), and the specificity of their interaction with enemies, plants should produce either quantitative or qualitative defenses (Feeny 1976, Rhoades & Cates 1976, Rhoades 1979). Plants that cannot “escape” from their natural enemies in space and time (i.e., trees) should produce quantitative defenses, which function as digestibility reducers on specialist herbivores (e.g., tannins produced by oaks, Feeny 1976). On the other hand, plants that do not have a predictable distribution (mainly herbaceous) should produce qualitative defenses (e.g., alkaloids and glucosinolates, Stamp 2003), which have a toxic effect on generalist or non-adapted consumers, but not on specialist (adapted) herbivores. For instance, in Arabidopsis, the larvae of the generalist lepidopteran Helicoverpa armigera, “cotton worm”, avoids feeding on rosette leaves with a high content of glucosinolates (Shroff et al. 2008), while other specialist lepidopteran species successfully feed on plants containing glucosinolates. Pieris rapae is a specialist herbivore of Brassicaceae and is biochemically adapted to the glucosinolate-myrosinase system of their host plants. After ingesting leaf tissue, P. rapae synthesizes a protein in the intestine which prevents the formation of isothiocyanates by reorienting the hydrolysis of glucosinolates toward the formation of nitriles which are excreted with feces. Some insects are capable not only of disabling the glucosinolato-myrosinase system, but can even use glucosinolates as a cue to locate their host plants (Walters 2011).
Counter-defensive strategies of phytophagous insects involve not only mechanisms that inhibit or circumvent plant defenses, but also behavioral strategies to inactivate the defenses of their host plants. For instance, the beetle Labidomera clivicollis (Coleoptera: Chrysomelidae) feeds on Asclepias syriaca, known to contain latex rich in cardenolides, which is pressurized in the veins of the plant. The beetle cuts lateral veins close to the central vein of the leaf, causing the latex to drain and lose pressure. Later, L. clivicollis feeds on the edges more distant to the leaf cuts. Vein cutting behavior has also been observed in other insect species that feed on Asclepias spp. (Malcolm 1991, Agrawal & Fishbein 2006, Agrawal et al. 2012) and on Blepharida species that feed on Bursera shclechtendalii (Becerra 1994). Likewise, constriction of petioles by larva of Erynnyis ello (Lepidoptera: Sphingidae) circumvents latex flow, allowing larva feed on Cnidoscolus urens but avoid the deleterious effects of sticky latex (Dillon et al. 1983). In addition, some herbivores use plant defense compounds to defend themselves. Insects that feed on Asclepias are able to “sequester” and store the cardenolides that bestowed defense on their host plants. Thus, cardenolides obtained through consumption of milkweeds subsequently protect insects from attack by predatory birds (Malcolm 1991).
Nonetheless, plants can also use other means (i.e., indirect defense) besides their own to defend themselves. One of the best known examples of indirect defense is the ant-plant association, also known as myrmecophily. Janzen (1966) documented one of the classic examples of a myrmecophytic association between the Acacia cornigera and its mutualistic ants Pseudomyrmex ferruginea. Ants inhabit the hollow spines (domacios) of the plant and feed on small globules rich in glycogen (Belt bodies) secreted by the apex of leaflets. The plant provides food and protection for the ants, while the ants provide defense to the plants by attacking herbivores that attempt to feed on the host. Additionally, in some cases, ants are able to remove plants growing on the myrmecophyte (Del Val & Dirzo 2004).
The above evidence of adaptive responses and counter-responses depicts the potential of reciprocal selective forces exerted by plants and herbivores to shape phenotypes of both linages and to influence evolutionary patterns. However, the extent to which plants and herbivores can co-evolve depends on at least four conditions: (1) plants and herbivores affecting each other’s fitness, (2) genetic variation for plant and herbivore traits that mediate their interaction, (3) response by each organism to selection imposed by the other (Janzen 1980, Strauss & Zangrel 2002) and (4) the relationship among defensive traits (e.g., trade-offs or synergisms; Rasmann & Agrawal 2009).
The “escape and radiation”: Host plants vs. herbivores. Ehrlich and Raven predicted that plant lineages diversify at a greater rate when the lineages are temporarily freed from herbivores, via novel defense. Circumvention of this defense by herbivores then allows herbivores lineages to radiate onto underused resources (Walters 2011). Secretory canals and latex are perhaps the best cases of novel defenses leading to radiation documented so far. Secretory canals are an effective defense against herbivorous insects and pathogens (Becerra 1994). They are highly convergent in angiosperms, occurring in 10 % of all species. Farrell et al. (1991) compared the diversities of lineages that possess (independently evolved) secretory canals with their sister groups and found that latex-bearing plant clades were significantly more species-rich than sister clades lacking secretory canals.
In a more recent study, Agrawal et al. (2009a) suggested that investment in defensive traits biases plant diversification. Agrawal and colleagues assessed defense-associated traits in milkweed, such as cardenolides and latex. They modeled the relationship between trait change and species diversification, while allowing rates of trait evolution to vary during radiation. The study shows that species-rich lineages experienced a proportionately greater decline in latex and cardenolides compared to species-poor lineages. Also, early in the radiation, the rate of trait change was faster. Nonetheless, while there is some evidence that evolution of particular defenses is coincident with adaptive radiation, mechanisms that allow coevolutionary dynamics to produce radiations are not fully understood. In this regard, much of the focus has been on explaining what causes host shift, and how these changes bias diversification.
Host shift has been clearly documented in several plant lineages. For instance, the prodoxid moth Greya politella (Lepidoptera: Prodoxidae) is a seed parasite of Lithophragma parviflorum. But, by extending its ecological range, populations of G. politella have colonized another saxifragous plant, the local endemic Heuchera grossulariifolia (Segraves et al. 1999). Heuchera grossulariifolia have both diploid and tetraploid variants and moths prefer to attack tetraploids. However, moths from remnant populations of the ancestral host, although they refused at first, were able to oviposit on the novel host, but were unable to differentiate between plants of different ploidy levels (Janz & Thompson 2002). This suggests that even when lacking adaptations to fit the new host, ancestral moths already had some ability to recognize and use the novel plant (Janz 2011). A preexisting condition that enables evolutionary change is a general explanation for evolutionary novelty proposed by West-Eberhard (2003). She introduced the term “phenotypic accommodation” to describe whether the first step for adaptive evolution is given by phenotypic plasticity. Either induced by mutation or by novel environmental factors, innovative morphological form comes from ancestral developmental responses, not from the novel inducing factor itself. Phenotypic accommodation is the result of adaptive developmental responses, and the novel morphologies that result are to some extent reflecting past functionality (West-Eberhard 2005). Host shifts may occur if the developmental pathways required to utilize a novel plant and the ancestral function overlap at some point. Thus, more plasticity will allow a wider range of such overlaps. Changes in feeding habits (e.g., polyphagy) should increase shifts by colonization of new hosts. Consequently, the diversification of host use through colonization appears to be closely linked to the evolution of host plant range (Janz 2011).
It has been suggested that insects can enlarge their host range in two fundamentally different ways: (1) through the accumulation of separate sets of specialized metabolic pathways (polyspecialism), and (2) through the evolution of generalized enzyme systems (true generalism) (Nylin & Janz 2009). The macroevolutionary consequences of these alternative paths of host-range expansion will be quite different. Unfortunately, the mechanistic bases and macroevolutionary patterns of host range evolution are still poorly understood (Janz 2011). The identification of the NSP (nitrile-specifying protein) glucosinolate detoxification gene as a key innovation involved in the host plant shift to Brassicales by butterflies in the Pirinae (Wheat et al. 2007), and the connection between diversification patterns and the role of a gene and genome duplications as a substrate for novel chemical defenses and molecular counteradaptations (Edger et al. 2015) are excellent examples of how genetic mechanisms can be linked to macroevolutionary patterns.
A particularly important contribution of molecular analysis is the incorporation of clade age, which has revealed congruent speciation patterns (but see Box 3) between host plant and herbivores (e.g., Farrell & Mitter 1998). Speciation patterns of host plants and herbivores may be synchronous or asynchronous (e.g., change of host plants). In an explicitly phylogenetic context, studies have shown that host use is highly conserved, ie. closely related herbivores feed on closely related plants (e.g., Funk et al. 1995, Becerra 1997, Janz & Nylin 1998). Host changes in less related plants occur only occasionally. But, when herbivores moved to a new host plant not closely related to the former host, both plant taxa usually resemble each other in chemical profiles (Becerra 1997). This indicates that in addition to relatedness, secondary metabolites are central to understanding diversification patterns. This supports the key role of (chemical) novelties as drivers of evolutionary dynamics.
Box 3. Parallel speciation
Close ecological relationships between plants and their consumers is not sufficient to explain speciation or phylogenetic relationships of host insects, nor demonstrate coevolution between the two groups. Even when plant-herbivore interactions may result in a phylogenetic pattern showing a taxonomic correspondence between both groups (i.e., phylogenetic congruence), this pattern may not always be attributed to a coevolutionary process, since two taxa can speciate in parallel and having no reciprocal responses between them. However, while the detection of reciprocal selection among interacting species is one of the strongest pieces of evidence supporting coevolution, the absence of reciprocal selection would not necessarily imply absence of coevolution. Thompson (1994, 2005) suggests that coevolution can be visualized as a mosaic at a geographic scale with intense reciprocal selection regions or “hot spots”, and non-reciprocal selection regions or “cold spots”. In this context, it cannot always be predicted what traits will be lost by extinctions, or extended throughout the geographic mosaic and eventually fixed (within a species). Though, in this scenario, patterns that may emerge at a phylogenetic scale are not very clear or not necessarily predictable.
To sum up this section, empirical evidence supports (1) the association of speciation in host plants and herbivores (Janz & Nylin 1998, Doorenweerd et al. 2015, Nakadai & Kawakita 2016), (2) asynchronous speciation patterns (host shifts) related to chemical compounds of host plants (Becerra 1997, Murphy 2004, Murphy & Feeny 2006), and in some cases (3) the association of diversification events or radiation rates with evolution of adaptive novelties related to plant defense (Edger et al. 2015). While most of the mechanistic basis of diversification and radiation patterns are not fully understood, recent advances in molecular genomic techniques (e.g., random introduction of mutations or functional copies within the genes, Chen et al. 2016) have substantially increased our knowledge of the genetic machinery behind macroevolutionary outcomes.
Overview of micro- and macroevolutionary perspectives for the study of plant defense. The notion that plant secondary metabolites are adaptations to defend themselves from herbivores, and that defense and counter-defense coevolution results in adaptive radiations of angiosperms and arthropod herbivores, contributed to the early conceptual understanding of plant defense (Agrawal 2006). This conceptual framework reflects the macro- and microevolutionary approaches derived not only from the rationale of the coevolutionary model, but from the way in which adaptive evolution is often assessed (Grandcolas & D´Haese 2003. Consequently, variation in plant defenses is frequently analyzed either within populations or across taxa. Although this analytical dichotomy has facilitated the study of adaptive traits, at the same time it can give rise to some conceptual problems: (1) the assumption that some microevolutionary events must scale phylogenetically (e.g., Ehrlich & Raven 1964), even when no mechanistic bases support this, (2) common phylogenetic patterns can be blindly assumed to be the result of processes occurring at the microevolutionary scale (e.g., natural selection), even when there is a lack of information on intraspecific dynamics or ignoring the effects of the time scale (see Erwin 2000). The taxonomic correspondence of lineages of plants and herbivores as exclusive evidence of coevolution, is a good example of this misconception of macroevolutionary patterns. Besides conceptual misinterpretation, the study of defensive traits at different scales has also imposed some methodological challenges. A common problem is the uneven development of tools to analyze data within populations or across phylogenies, which also leads to a bias in the availability of empirical evidence that may support particular hypotheses.
The microevolutionary approach based on quantitative genetic methods to measure natural selection, heritability and genetic variance within species, has dominated the study of plant defense. Nonetheless, over the past twenty years the development of rigorous tests and comparative methods have allowed the testing of comparative hypotheses, and elucidation of major phylogenetic patterns (e.g., Martins & Hansen 1997, Pagel 1999). Yet, the limited access to biological material of several species and the lack of molecular phylogenies for non-model species still leads to methodological challenges for the study of plant defense and biological interactions. Hence, there are far fewer phylogenetic studies examining the defense of plants than the large amount of work at microevolutionary scale. Comparative analysis however, has proved in a few years to be a useful tool to better understand the evolutionary dynamic of traits involved in the interaction of plants and herbivores. For instance, phylogenetic studies have shown that covariation between defensive traits is adaptive, and often shaped by the herbivore community (e.g., Agrawal & Fishbein 2006, Agrawal et al. 2009b). Analyzing the dynamics of covariation between plant attributes across phylogeny could help to recognize not only the adaptive value of defensive traits, but also of their associations themselves. In a recent study, Kariñho-Betancourt et al. (2015) showed that correlations among defensive traits change according to plant ontogeny. The authors analyzed the correlation between five tropane-alkaloids and leaf trichomes across the genus Datura at two ontogenetic stages (juvenile vs. reproductive). They found significant phylogenetic correlations among alkaloids and trichomes occurring only at the juvenile stage. These patterns show how “sensitive” the relationship among plant attributes is, to intrinsic and/or extrinsic changes over the plant’s life. This study suggests not only that the associations are adaptive, but also that the plasticity of relationships between traits may have an adaptive value.
One of the major contributions of the comparative approach to the study of plant defense is that it exposes the common mechanisms underlying species interactions. For instance, this approach has contributed to the identification of jasmonic acid as the hormonal regulator of plants’ induced responses to herbivores attack and other biotic and abiotic stimuli. Jasmonic acid is involved in the production of diverse plant defenses such as glucosinolates, oxidative enzymes, alkaloids and trichomes, among others (Traw & Bergelson 2003, Heil et al. 2004, Howe 2004). Nonetheless, to detail the biotic or abiotic contexts in which such chemical compounds are expressed and corroborate the current (defensive) functionality attributed to them, targeted microevolutionary studies are needed. Hence, a comprehensive approach including both the micro- and macroevolutionary perspectives to study plant defense would help to construct a rich understanding of adaptive evolution.
Concluding Remarks. The interdisciplinary study of the interaction of plants with arthropod herbivores mediated by secondary compounds, has begun to unravel co-diversification patterns (Janz 2011). From this, trends can be outlined: (1) plants and herbivores constitute a selective context for each other, (2) plant secondary metabolites are adaptations to interact with herbivores, (3) the ecological role of secondary metabolites is tied to the degree of specialization of the interaction, (4) insect host shifts are constrained by host plant chemistry, (5) novel plant defenses, resulting from coevolution, are associated with an increase in diversification and radiation rates, and (6) gene and genome duplications are common mechanisms from which evolutionary novelty is achieved.
The idea of plant secondary compounds as waste products from over 50 years ago now seems like a faint memory. Much of the progress made during the last decades has come from the development and application of comparative analysis and molecular techniques. However, given how extraordinarily diverse the interaction of plants and herbivores is, there is still much to unravel. In particular, much work needs to be done to disentangle the mechanisms behind speciation and diversification, and the machinery that affords or precludes variation within or across species. For this, worked-out case studies integrated with comparative approaches, and the use of modern molecular techniques may be a fruitful avenue of research.











 text new page (beta)
text new page (beta)