Introduction
Genetic variation is considered to be the basic level of biological diversity (McNeely et al. 1990) and is essential for the adaptation and survival of individuals, the viability of populations, and the ability of species to adapt to environmental changes (Frankham et al. 2010). Particularly since 1970, genetic variation started being regarded as a key element of conservation (Frankel 1974). On the other hand, the acknowledgment of the impact of human activities on the environment in the 1980s — mainly fragmentation and habitat loss — gave rise to landscape ecology as the discipline that investigates the interactions between spatial heterogeneity and ecological processes (Turner 2005). In 2003, the conceptual and methodological development of both disciplines laid the foundations for the recognition of Landscape Genetics (LG) as a discipline that evaluates the impact of environmental heterogeneity and the landscape elements on the variation and genetic structure of individuals and populations (Manel et al. 2003). LG integrates the concepts and tools of population genetics, landscape ecology, and spatial statistics, to quantify the effects of the landscape matrix (composition, configuration, and quality) on microevolutionary processes such as gene flow, drift and selection, based on neutral or adaptive genetic variation (Manel et al. 2003; Holderegger and Wagner 2006; Storfer et al. 2007; Balkenhol et al. 2015).
The impact of LG in the scientific community has led to a marked increase in the number of publications related to the topic, from 3 to 60 articles per year, over 10 years (Storfer et al. 2010). A variety of analytical and methodological approaches have emerged, including the stages that a LG study should follow (Garrido-Garduño and Vázquez-Domínguez 2013; Hall and Beissinger 2014), the consideration of temporal and spatial scales (Anderson et al. 2010), the sampling design and selection of molecular markers (Landguth et al. 2012), statistical analyses (Balkenhol et al. 2009a), and certain limitations and perspectives of LG (Balkenhol et al. 2009b; Richardson et al. 2016). In this sense, Storfer et al. (2010) analyzed empirical studies published up to that date; from their results, five points stand out: 1) there is a taxonomic bias toward vertebrates; 2) most works have been conducted in America; 3) forests are the most studied habitats; 4) the topic most frequently addressed is the identification of barriers affecting gene flow; and 5) deserts are scarcely represented ecosystems, with 3 % of LG studies. Other revisions also point out that vertebrates are the dominant taxonomic group in LG studies at the global level (Garrido-Garduño and Vázquez-Domínguez 2013; Dyer 2015); of these, mammals have prevailed (Montgelard et al. 2014). It should be noted that although America is the region most represented in LG studies, there is no information currently available identifying the main topics of research, analytical methods used, and environments studied from an LG perspective.
Deserts are one of the Earth’s more widespread environments, occupying approximately one-third of the Earth’s surface (Schimel 2010). The environmental characteristics of deserts, such as high temperatures and low rainfall, have favored a variety of microhabitats that, in addition to hosting a large number of taxonomic groups, many of them endemic, also confer temporal and spatial heterogeneity (Whitford 2002; WWF 2019). Temporal heterogeneity emerges from highly variable environmental conditions throughout the day, between seasons of the year, or between years (Polis 1991); for its part, spatial heterogeneity influences species composition, distribution, and abundance (Whitford 2002; Ludwig et al. 2005). Thus, heterogeneity renders deserts ideal systems for hypothesis testing within the LG framework (Challenger and Soberón 2008). For instance, in the Chihuahuan desert, vegetation tends to be distributed in small patches (Grünberger 2004), producing a heterogeneous matrix that allows evaluating structural and functional connectivity (Manel and Holderegger 2013).
It is worth stressing that in the current context of global climate change, it is predicted that change rates toward warmer and more arid environments will rise in deserts relative to other regions. This, coupled with the transformation and loss of natural habitats in desert ecosystems (Mittermeier et al. 2003; Zeng and Yoon 2009; WWF 2019), have supported that deserts be presently considered as vulnerable regions, particularly the deserts of North America (Bachelet et al. 2016; WWF 2019). Therefore, due to the overall scarce representation of deserts in the literature of landscape genetics (Storfer et al. 2010), we deem it essential to determine the state of the art of LG research in American deserts. Its relevance lies in the fact that the persistence of species inhabiting desert ecosystems depends on the dispersal ability of individuals and the movement of genes within and between their populations across the landscape (Scribner et al. 2005; Reding et al. 2013).
Based on the above arguments, in the present work we conducted a review of the scientific literature aiming to 1) determine the most studied groups of mammals; 2) determine the representativeness of desert ecosystems; 3) describe the research questions most frequently addressed and the methods and analysis used; and 4) summarize the main factors of the landscape and the environment associated with genetic diversity and structure of the mammals of America.
Materials and Methods
Literature review. We surveyed articles about landscape genetics (LG) with mammals in ecosystems of America published between 2003 — when the term was first coined — and May 2019. We used the Web of Science website (http://apps.webofknowledge.com) based on different combinations of terms as keywords: ‘landscape genetics’, ‘functional connectivity’ and ‘mammals’; ‘landscape genetics’, ‘mammals’ and ‘desert’; ‘landscape genetics’, ‘mammals’, ‘desert’ and ‘America’; ‘functional connectivity’, ‘mammals’ and ‘desert’. This literature search method has proved to be efficient in different review works (e. g., Storfer et al. 2010; Dyer 2015; Rico 2019), which also allows a systematic and repeatable analysis, although it certainly may exclude some works.
Literature validation and analysis. We conducted a detailed revision of the works obtained to the last screen (revision of the title, abstract, and methods), aiming to eliminate duplicated works and confirm that studies matched the search criteria (mammals, America, and desert). We considered only works that reported empirical data and that strictly corresponded to genetic landscape analysis, i. e., including at least one landscape variable and evaluating its relationship with genetic patterns. We collected information on species, authors, year of publication, environments (considering the most represented environment in the study area according to each work), research questions (classified by ‘type’), and statistical-spatial LG analysis used. The research questions were classified according to six types: ‘connectivity’, ‘structure’, ‘gene flow’, ‘comparative analysis’, ‘association analysis’ and ‘adaptation’.
Results
Taxonomy, geographic region, and diversity of deserts. The results obtained for each combination of terms and search screened were as follows: ‘landscape genetics’, ‘functional connectivity’ and ‘mammals’ (n = 85 works); ‘landscape genetics’, ‘mammals’ and ‘desert’ (n = 78); ‘landscape genetics’, ‘mammals’, ‘desert’ and ‘America’ (n = 38); ‘functional connectivity’, ‘mammals’ and ‘desert’ (n = 10). After a thorough validation of the literature (elimination of duplicates, inclusion of works with empirical data only involving the analysis of landscape variables), a total of 36 publications were obtained (Table 1). We identified six orders of mammals, of which Rodentia was the most represented taxon (n = 20; Table 1). The main geographic region studied was North America (n = 25), mostly in the United States of America (n = 19). For South America (n = 10), we identified works conducted in Brazil (n = 3), Argentina (n = 6), and Uruguay (n = 1). We recorded one study that covered countries of North America (Mexico) and Central America (Belize, Costa Rica, Guatemala, and Honduras), which was counted separately. The environments involved included forests (n = 6), urban areas (n = 6), cropland (n = 4), mountains and rivers (n = 5), sand dunes (n = 5), and shrub steppe (n = 2), while desert ecosystems were addressed in eight studies.
Table 1 Summary of landscape genetics studies with mammals analyzed in this review.
| ORDER/Species | F | Region | Environment/landscape | Research question | Type of question | Statistical-spatial analysis | Genetic structure and diversity drivers |
|---|---|---|---|---|---|---|---|
| DIDELPHIMORPHIA | |||||||
| Marmosops incanus | 1 | Atlantic Plain, Brazil | Forests with different degrees of fragmentation | Comparative landscape genetics Response of genetic diversity to fragmentation | Comparative analysis | ANOVA, regression models, and Mantel tests | Amount of available habitat across the landscape (% patch cover) |
| PRIMATES | |||||||
| Leontopithecus rosalia | 2 | Uniao Biological Reserve, Brazil | Fragmented Atlantic forest | Evaluate the effect of the landscape in gene flow | Gene flow | Spatial autocorrelation, kinship indices, and generalized linear models | The spatial configuration of vegetation cover affects the dispersal of individuals |
| LAGOMORPHA | |||||||
| Brachylagus idahoensis | 3 | Wyoming, Estados Unidos | Shrub steppe | Evaluate whether the spatial genetic structure pattern is due to isolation by distance or by barriers | Structure | Analysis of isolation by distance at individual and population levels | Geographic distance, with a road as a likely barrier |
| Ochotona princeps | 4 | Oregon, Estados Unidos | Mountains and rivers | Identify the factors that limit or facilitate gene flow | Gene flow | Genetic distance, resistance distances, simple and partial Mantel tests, simulations | Topographic complexity is the main driver of gene flow |
| Sylvilagus transitionalis | 5 | Eastern United States | Fragmented habitat / Urban zone | Assess connectivity | Connectivity | Resistance matrix, partial Mantel tests, Least-cost path and mixed-effect models | Linear anthropogenic constructions and shrub habitats (effective distance) |
| RODENTIA | |||||||
| Calomys venustus | 6 | Córdoba, Argentina | Farming areas with roads | Assess spatial and temporal genetic structures | Structure | Spatial autocorrelation analysis, Mantel and partial Mantel tests. Correlation between genetic and “geographic” distances | Geographic distance only |
| Ctenomys “chasiquensis” | 7 | Las Pampas, Argentina | Sand dunes and cropland | Evaluate the environmental factors that shape population structure and those that promote the connectivity between populations | Structure | AMOVA, simple and partial Mantel tests, spatial principal component analysis (sPCA), generalized linear models | Plant cover (NDVI) and Longitude promote gene flow between populations |
| Ctenomys lami | 8 | Coxilha Lombas, Brasil | Matrix of rivers, lagoons and cropland | Assess the spatial genetic structure | Structure | Correlation between genetic and geographic distances, Mantel tests, assignment analysis, AMOVA | Geographic distance, probably due to the species limited dispersal |
| Ctenomys porteousi | 9 | Buenos Aires, Argentina | Cultivation and farming areas | Evaluate the effect of landscape configuration on genetic structure and connectivity. Migration rates | Connectivity | Connectivity between habitat patches. Correlation between genetic and geographic distances. Mantel and partial Mantel tests, generalized linear model. | Amount of available habitat across the landscape and distance between good-quality patches. |
| Ctenomys rionegrensis | 10 | Río Negro, Uruguay | Dune and river systems | Evaluate the geographic factors that shape population structure | Structure | Mixed generalized models | Elevation |
| Ctenomys sp. | 11 | Corrientes, northeast Argentina | Flood-prone area including lagoons, marshes, and cropland | Evaluate the geographic factors that shape population structure | Structure | Analysis of structure (GESTE), species distribution models and linear models | The presence of well-drained sandy soils and temperature are the drivers for the distribution and differentiation of populations |
| * Dipodomys merriami | 12 | Mapimí, Durango, México | Chihuahuandesert | Evaluate the landscape features that limit or facilitate gene flow | Gene flow | Linear mixed-effect models from resistance surfaces and model evaluation using AIC | Effective distance (NDVI) best explains gene flow patterns, which is favored in areas with vegetation cover |
| * Dipodomys spectabilis | 13 | Nuevo México, Estados Unidos | Chihuahuandesert | Evaluate the presence of a founder effect on recolonized sites based on genetic diversity, size, and connectivity between sites | Connectivity | Correlation between genetic and geographic distances. Mantel test, mixed generalized models | Dispersal characteristics associated with population density |
| Hydrochoerus hydrochaeris | 14 | Basins of Venezuela, Paraguay, and Argentina | River systems | Assess spatial genetic structure | Structure | AMOVA. Principal Coordinate Analysis and Canonical Correspondence. Resistance distance | Rivers determine the structure pattern |
| Lagidium viscacia | 15 | Neuquén, Argentina | Steppe and mountains | Evaluate functional connectivity | Connectivity | Correlation between genetic, geographic and cost distances. Mantel tests | Functional connectivity is influenced by landscape geology |
| Liomys pictus | 16 | Western Mexico | Tropical deciduous forest | Evaluate the effect of landscape elements on genetic structure and gene flow. | Gene flow | Environmental domains. Wombsoft, Barrier. Correlation between genetic and geographic distances. Effective distance: Least-cost path and circuit theory. Mantel and partial Mantel tests | Effective distance, precipitation and streams |
| Microtus californicus | 17 | Jasper Ridge Reserve, California, United States | Grasslands and oak forest | Evaluate gene flow | Gene flow | Principal component analysis, Mantel tests | Only geographic distance, due to the ecological characteristics. |
| Ondatra zibethicus | 18 | Sudbury, Ontario, Canada | Watersheds | Evaluate the effect of landscape characteristics on structure and connectivity. | Connectivity | Spatial autocorrelation by sex. Assignment analysis. Correlation between genetic, linear and resistance distances (least-cost path). PATHMATRIX. Partial Mantel tests | Roads and anthropogenic elements seem to facilitate the movement of organisms |
| Peromyscus leucopus | 19 | New York, United States | Urban zone | Evaluate the landscape characteristics that foster connectivity between populations in an urban environment | Connectivity | Migration rates (Nm, BayesAss and Migrate-n). Linear, effective (least-cost) and resistance distances. Mantel and partial Mantel tests | Effective distance (based on vegetation cover) |
| P. leucopus | 20 | Montérégie, Quebec, Canada | Farming land with rivers and roads | Evaluate the effect of the landscape characteristics on genetic structure and connectivity. | Connectivity | Correlation between genetic and ecological distances. Mantel tests. Multiple regression analysis between ecological and linear distances. Connectivity between patches with resistance distance | Forest fragments facilitate the movement of individuals |
| Rattus norvegicus | 21 | Baltimore, Maryland, United States | Urban zone | Characterize the genetic structure and evaluate gene flow | Gene flow | Genetic distance, kinship relations, and Mantel tests | Habitat fragmentation (urban area); the ecology of organisms contribute to homogenize diversity and genetic structure |
| R. norvegicus | 22 | New York, United States | Urban zone | Explore spatial genetic structure patterns | Structure | Spatial autocorrelation, Mantel tests, PCA, sPCA, estimated effective migration surfaces (EEMS), population structure (fineSTRUCTURE) | Closely related individuals and ecological characteristics of the species |
| Tamias striatus | 23 | south Quebec and Ontario, Canada | Forested areas, rivers, and urban areas | Explore the geographic factors that shape population structure; sex-biased dispersal | Structure | Correlation between genetic and geographic distances Mantel test. Identification of barriers (Barrier). AMOVA | The river is the main barrier, as well as sex-biased dispersal (males) |
| T. striatus | 24 | Indiana, United States | Patches of forest and farmland | Evaluate functional connectivity | Connectivity | Assignment analysis. Correlation between genetic and geographic distances. Mantel test. Coverage distances. Regression models | Vegetation cover promotes gene flow |
| Zapus trinotatus | 25 | Olympic Peninsula, Washington, United States | River systems, presence of mountains and forest | Assess connectivity in among rivers using three environmental distances. Migration rate | Connectivity | Spatial autocorrelation analysis. Mantel test. Estimation of migration rates | Effective distance (topographic and riparian landscape features) and limited species dispersal |
| CARNIVORA | |||||||
| * Bassariscus astutus | 26 | Southwest United States | Desert and mountainsChihuahuan | Assess connectivity patterns associated with genetic structure: IBD, IBR, IBE | Connectivity | Partial Mantel tests. Discriminant function analysis. ANOVAs | Environmental characteristics: elevation, slope, and vegetation type (IBE-effective distance) |
| Canis lupus | 27 | Rocky Mountains, Canada | Mountains | Evaluate gene flow between herds considering the landscape characteristics | Gene flow | Regression between genetic and geographic distances (resistance model and coverage distance). Multiple regressions of distance matrices. Partial Mantel tests | Effective distance (based on vegetation cover) |
| Martes pennanti | 28 | Northeast United States | Mountains and rivers | Characterize genetic structure and its association with landscape features and human disturbance | Structure | Correlation between genetic, geographic and barrier distances. Mantel and partial Mantel tests. Recent migration rates | Orographic and hydrological characteristics (i.e. river and great lakes) |
| Panthera onca | 29 | In Mexico: Sierra Mixe, Oaxaca, and Sierra de Abra-Tanchipa, San Luis Potosí. In Central America: Belize, Costa Rica, Guatemala, Honduras | Tropical deciduous forest and medium tropical forest | Evaluate genetic structure at different spatial scales | Structure | Analysis of population structure, PCA, AMOVA, spatial autocorrelation, and Mantel tests. | Geographic distance, probably due to habitat fragmentation |
| * Ursus americanus | 30 | Arizona, United States | Mountains andSonoran and Chihuahuan deserts | Evaluate connectivity and identify potential corridors | Connectivity | Analysis of occupation based on landscape characteristics (e.g. coverage). Habitat availability model. Resistance layers and corridor modeling | Fragmentation/barrier associated with the construction of the border wall |
| U. americanus | 31 | Michigan, United States | Cultivation and farming areas | Evaluate the effect of landscape changes on spatial genetic structure through time. | Structure | Simple and partial Mantel tests, resistance distances, FRAGSTATS, autoregressive spatial model, selection of models based on AIC | Vegetation cover showed a better relationship with genetic distance, while variables associated with environmental heterogeneity better predicted the genetic change over time |
| ARTIODACTYLA | |||||||
| Odocoileus hemionus | 32 | Southern California, United States | Mountains and urban areas | Assess connectivity | Connectivity | Genetic structure (STRUCTURE and DAPC), genetic distances between individuals, habitat accumulated cost distance (HAC), least-cost path, generalized linear mixed-effect models | Highways restrain the connectivity between populations and individuals |
| * Ovis canadensis nelsoni | 33 | Southeast California, United States | Mojave and Sonorandeserts | Assess connectivity between populations, considering the effect of slope and anthropogenic presence | Connectivity | Migration rates. Correlation between genetic and geographic distances. Least-cost path. Partial Mantel test | Effective distance (based on topography) |
| * O. c. nelsoni | 34 | Southwest United States | Mojavedesert | Combine connectivity by landscape resistance models and network theory to prioritize patches or corridors for conservation purposes | Connectivity | Effective resistance distance (least-cost path). Partial Mantel tests. Genetic and demographic network models. Correlations | Effective distance (based on maximum dispersal) |
| * O. c. nelsoni | 35 | Southwest United States | Mojavedesert | Implementing NDVI as a predictor of food quality and genetic diversity | Association | Linear quadratic regression models to measure the association between NDVI and genetic diversity (response variables). Connectivity metrics as an additional predictor variable | Patches with high NDVI values |
| * O. c. nelsoni | 36 | Southwest United States | Mojavedesert | Explore the effects of landscape on the spacing of adaptive genetic variation | Adaptation | Linear regression models from resistance (least-cost) surfaces and simulations | Effective distance (terrain slope, water bodies,and roads determine gene flow) |
Abbreviations: IBD= isolation by distance (Isolation by Distance), IBR= isolation by barrier (Isolation by Barrier), IBE= isolation by ambient (Isolation by Environment); ANOVA = Analysis of variance; AMOVA= analysis of molecular variance; PCA = Principal component analysis.
*= study explicitly conducted in a desert.
F= Source. 1: Balkenhol et al. (2013); 2: Moraes et al. (2018); 3: Thimmayya and Buskirk (2012); 4: Castillo et al. (2014); 5: Amaral et al. (2016); 6: Chiappero et al. (2016); 7: Mora et al. (2017); 8: Lopes and De Freitas (2012); 9: Mapelli et al. (2012); 10: Kittlein and Gaggiotti (2008); 11: Gómez Fernández et al. (2016); 12: Flores-Manzanero et al. (2019); 13: Cosentino et al. (2015); 14: Byrne et al. (2015); 15: Walker et al. (2007); 16: Garrido-Garduño et al. (2016); 17: Adams and Hadly (2010); 18: Laurence et al. (2013); 19: Munshi-South (2012); 20: Marrotte et al. (2014); 21: Gardner-Santana et al. (2009); 22: Combs et al. (2018); 23: Chambers and Garant (2010); 24: Anderson et al. (2015); 25: Vignieri (2005); 26: Lonsinger et al. (2015); 27: Cullingham et al. (2016); 28: Hapeman et al. (2011); 29: Wultsch et al. (2016); 30: Atwood et al. (2011); 31: Draheim et al. (2018); 32: Fraser et al. (2019); 33: Epps et al. (2007); 34: Creech et al. (2014a); 35: Creech et al. (2014b); 36: Creech et al. (2017)
Specifically for deserts, Artiodactyla was the order most represented (n = 4), although with a single species (the desert bighorn sheep Ovis canadensis nelsoni), followed by the orders Carnivora (Ursus americanus and Bassariscus astutus) and Rodentia (Dipodomys spectabilis and Dipodomys merriami), with two works each. The deserts of North America (Mojave, n = 3; Chihuahuan, n = 3; Sonoran n = 2) were the only arid environments where LG studies have been carried out (Table 1). No studies were found addressing the deserts of South America (Atacama and Patagonia). For Mexico, we found only three LG works, two in the tropical deciduous forest of Oaxaca and San Luis Potosí, and the other one the coast of Jalisco; only a single study was conducted in the desert (Chihuahuan; Table 1).
Research questions and statistical-spatial methods. The classification of the research questions addressed in all the publications reviewed (North America) revealed that those dealing with ‘connectivity’ and ‘structure’ were the most common ones (n = 14 and 12, respectively), followed by ‘gene flow’ (n = 7). The ‘comparative’, ‘association’ and ‘adaptation’ approaches were represented by one work each. To note, the ‘association’ and the ‘adaptation’ approaches were unique to desert environments (Table 1).
All studies analyzed genetic aspects that included assessment of deviations from the Hardy-Weinberg equilibrium and linkage disequilibrium, as well as genetic diversity measures by estimating allelic richness and observed and expected heterozygosity. Four works performed allocation and structure analyses to define genetic groups and to detect migrants (Figure 1).
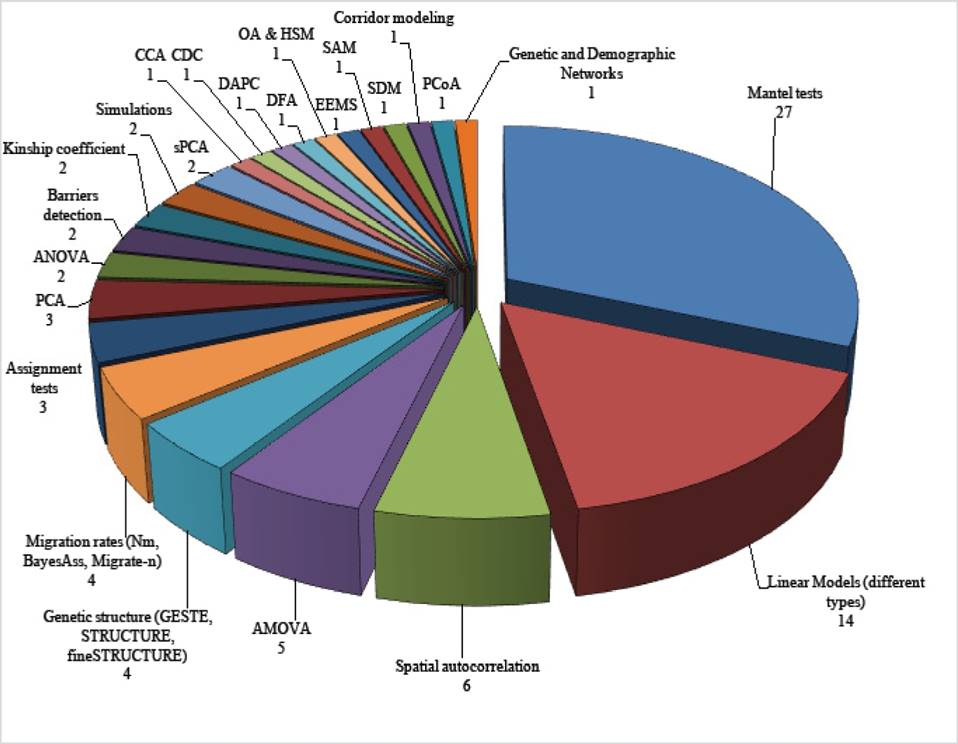
Figure 1 Analytical methods used in landscape genetics studies with mammals in North America. Numbers indicate the number of studies that used each method; all works used more than two methods. Abbreviations: ANOVA = Analysis of variance; AMOVA = Analysis of molecular variance; CEC = Canonical correspondence analysis; CDC = Climatic domain classification; DAPC = Discriminant principal component analysis; DFA = Discriminant functional analysis; EEMS = Estimated effective migration rates; OA and HSM = Occupancy analyses and Habitat suitability model; PCA = Principal component analysis; PCoA = Principal coordinate analysis; sPCA = Spatial principal component analysis; SAM = Spatial autoregressive modeling; SDM = Species distribution models.
Regarding spatial data handling and recording (landscape and environmental variables), all studies used a Geographic Information System (GIS) to represent the study area. In addition, five works estimated the Normalized Difference Vegetation Index (NDVI) to obtain vegetation cover data. A study selected sampling sites based on the method of environmental domains (Figure 1). In the remaining works, the landscape was characterized through a classification based on the literature reported for the area, or from repositories of specific environmental information.
In all the studies at least two analytical methods were used for assessing the genetics-landscape relationship. The methods most commonly utilized were the Mantel and partial Mantel tests, followed by linear regression models and their variants (n = 14; Figure 1). Some works addressing ‘connectivity’ and ‘gene flow’ questions constructed resistance layers to quantify the effects of landscape through the estimation of effective distances based on least-cost path algorithms (n = 6), resistance distance (estimated based on circuits theory; n = 6), and both (n = 4). One work used species distribution models (SDM) for the construction of these resistance layers (Figure 1).
Landscape factors that determine the genetic structure and diversity. The results obtained for America show that the environmental and landscape characteristics, represented as effective distances, are the ones that best explained genetic structure and diversity patterns in most works (Figure 2a), followed by geographic distance (i. e., with a pattern of isolation by distance). The main landscape variables included topography, vegetation cover (evaluated as NDVI and extent of available habitat in the landscape), rivers, and water bodies (Figure 2b). Additional relevant factors were anthropogenic constructions (highways and roads) and the ecological characteristics of species, such as dispersal capacity and population density. In particular in deserts, the factors most frequently associated with genetic structure and diversity were vegetation cover, dispersal, and anthropogenic constructions (Figure 2b).
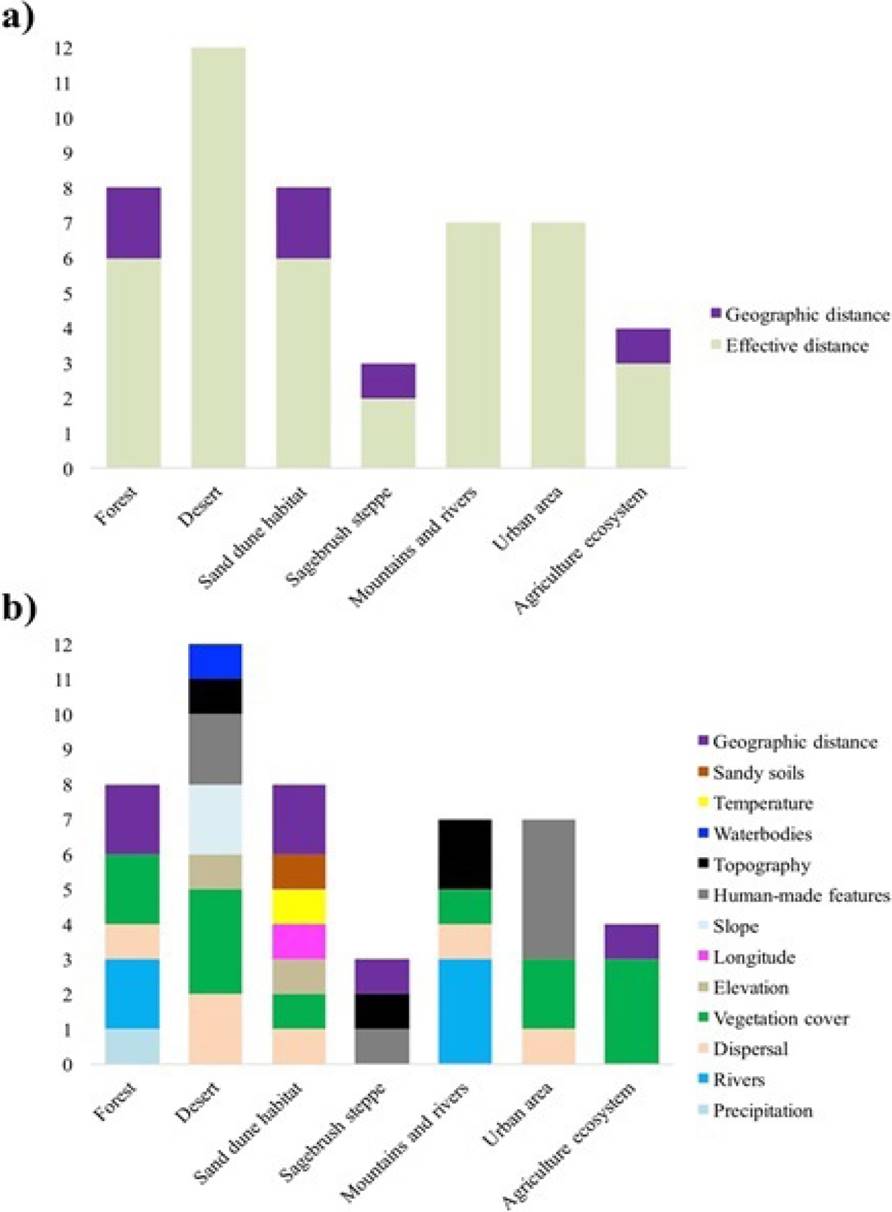
Figure 2 Factors that determine the genetic structure and diversity of mammals, expressed as accumulated frequencies in each of the environments analyzed. “Vegetation cover” includes the different approaches used to assess that characteristic (NDVI, habitat patches), as well as for “anthropogenic constructions” (roads, highways). a) Factors in terms of geographic and effective distance; b) environmental and landscape variables that constitute effective distances in each environment. “Longitude” refers to the geographic position in the coordinate system.
Discussion
Since the term was first coined to the steady construction of the theoretical, methodological, and analytical frameworks (Manel et al. 2003; Holderegger and Wagner 2006; Balkenhol et al. 2015), landscape genetics (LG) has been broadly accepted in the scientific community to address questions related to the effect of the environmental variables in the genetic structure and variation of natural populations (Storfer et al. 2010; Garrido-Garduño and Vázquez-Domínguez 2013; Dyer 2015). Mammals comprise 6,399 species worldwide (of the 6,495 recognized species, 96 have become extinct); of these, 697 are located in the Nearctic region, which covers North America (Burgin et al. 2018). Our results show that this region is the best represented in LG studies with mammals, covering a great diversity of ecosystems. A study that encompasses North America and Central America is worth mentioning (Wultsch et al. 2016), and could be considered as one of the first addressing LG for this region. In addition, we found scarce research efforts done (and published) on landscape genetics involving the mammals of Mexico and South America, as well as in desert ecosystems.
Rodents and carnivores, models in landscape genetics in North America. Rodentia is considered the largest order, with 2,409 species that account for approximately 44 % of the diversity of mammals worldwide (Wilson et al. 2017). The characteristics of rodents, namely high species richness, small size (most species), limited dispersal capabilities, and diverse life stories, make them an ideal taxonomic group to address questions at the landscape scale, together with genetics and ecology. It is striking, however, that it was only recently that the qualities of this group were recognized within the context of landscape studies (Waits et al. 2015).
Studies involving rodents in America have been conducted mainly in areas with anthropogenic impact, including urban areas (Gardner-Santana et al. 2009; Chambers and Garant 2010; Mapelli et al. 2012; Munshi-South 2012; Marrotte et al. 2014; Anderson et al. 2015; Chiappero et al. 2016; Combs et al. 2018). On the other hand, we found only two works that were conducted with rodents in desert environments, specifically kangaroo rats in the Chihuahuan desert. In the first study, Cosentino et al. (2015) evaluated the population genetics of D. spectabilis from a spatial perspective, considering the configuration of the landscape matrix; they found that genetic differentiation patterns of its populations are determined by biological aspects of the species, specifically the dispersal capacity and population densities. In the second work, Flores-Manzanero et al. (2019) found higher gene flow in a population of D. merriami along areas with shrub vegetation, which in turn is associated with the construction of burrows and as a food source (seeds); accordingly, based on a LG approach these authors found a relationship between genetic patterns and ecological processes.
Carnivores were the second most represented order in landscape genetics studies in America, encompassing both medium-sized species such as the fisher (Martes pennanti; Hapeman et al. 2011) and the “cacomixtle” (ringtail, Bassariscus astutus; Lonsinger et al. 2015), as well as large species, including the gray wolf (Canis lupus; Cullingham et al. 2016), the black bear (Ursus americanus; Atwood et al. 2011; Draheim et al. 2018), and the jaguar (Panthera onca; Wultsch et al. 2016). Notably, carnivores are recognized as the most vulnerable to extinction group of mammals, due to their biological characteristics and anthropogenic impacts (Cardillo et al. 2004). The few studies identified in this review highlight the importance of conducting genetic landscape research to explore, among others, the effect of landscape elements (mountains, rivers) and anthropogenic features (habitat fragmentation) on the diversity and genetic structure of this group.
Evaluation of connectivity. Landscape genetics has focused primarily on terrestrial organisms (Storfer et al. 2010; Garrido-Garduño and Vázquez-Domínguez 2013; Dyer 2015), where the development of tools, like geographic information systems (GIS), has allowed a more real representation of terrestrial landscapes (Waits et al. 2015). In this review, we found that one of the LG questions most frequently addressed globally (America) was the connectivity between populations, and all studies evaluating this topic used GISs to represent the landscape. These studies also used different analytical methods to relate the landscape with the genetic structure of populations, including the correlation between genetic distances and different geographic distances, since just the linear (Euclidean) distance does not represent the true distance between populations (Vignieri 2005). The so-called effective distance or functional distance represent ecological measures of distance commonly used in LG studies, the most used being the least-cost path and the resistance distance (Storfer et al. 2007; McRae et al. 2008). Also, the Mantel and partial Mantel tests were the approaches most frequently used to evaluate the correlation between these distances. Although the use of Mantel tests has been questioned (see Guillot and Rousset 2013), they are considered appropriate methods when setting hypotheses that explicitly involve distance (Legendre and Fortin 2010; Legendre et al. 2015), while also favoured as models that are easy to interpret and serve as starting points for the parameterization of resistance matrices (Storfer et al. 2010). For example, Munshi-South (2012) evaluated the connectivity between populations of the white-footed mouse (Peromyscus leucopus) in an urban area using estimates of migration rates and correlating these with cost and resistance distances, supported on the resolving power of GISs and Mantel and partial Mantel tests. Also, in an environment with anthropogenic impact, Amaral et al. (2016) assessed the connectivity between populations of the gray rabbit (Sylvilagus transitionalis) from cost and resistance distances, which were optimized using correlations through partial Mantel tests. Likewise, Draheim et al. (2018) implemented simple and partial Mantel tests to correlate genetic distances between paired individuals with geographic (linear) and resistance distances, to evaluate whether changes in the landscape over time influenced the spatial genetic structure of the black bear (Ursus americanus).
An aspect worth mentioning is the development of additional methods to assess connectivity in terms of the relationship between effective and genetic distances, like the generalized linear models, which prevailed in studies with rodents (Kittlein and Gaggiotti 2008; Mapelli et al. 2012; Marrotte et al. 2014; Cosentino et al. 2015). We should also highlight the use of species distribution models for the construction of connectivity hypotheses in landscape genetics, as these combine presence data (localities where the species has been recorded) and climate data associated with that locality, also considering vegetation cover, topography, and other environmental variables (Rolland et al. 2015). For instance, Gómez Fernández et al. (2016) modeled the distribution of the tuco-tuco (Ctenomys sp.), taking into consideration the environmental variables available for the flooded area where it thrives, which allowed determining that permeable sandy soils and temperature are the factors significantly associated with its population genetic structure. Notably, the use of species distribution models in LG studies is restrained by the geographic scale, since most of the variables used for constructing these models are available at large scales (for example, the WorldClim layers have 1 km2 resolution; Hijmans et al. 2005) and, therefore, their use is restricted to considering large areas.
Finally, we highlight the construction of genetic and demographic networks from resistance matrices. As an example, Creech et al. (2014a) combined resistance models with the network theory and evaluated their correlation with partial Mantel tests to identify patches and corridors that facilitate the connectivity between populations of the bighorn sheep (Ovis canadensis nelsoni). Thus, this analytical approach can be useful for identifying corridors, particularly in conservation studies.
Not surprisingly, connectivity is one of the topics most commonly addressed in landscape genetics, particularly under the current context of global changes that currently affect (and will continue to affect) the habitat of multiple species (Manel and Holderegger 2013).
Genomics, gene flow, and local adaptation. The work in landscape genetics has been based largely on the use of neutral genetic markers, such as microsatellite loci. However, given the accelerated transformation of natural environments, it is increasingly important to be able to assess the patterns derived from adaptive genetic variation, especially because there lies the potential of species to respond to these changes (Manel et al. 2003; Balkenhol et al. 2015). In this sense, the use of molecular markers such as SNPs (Single Nucleotide Polymorphisms) has made it possible to multiply the number of loci by the thousands, increasing the statistical power of landscape genetic analyses (Combs et al. 2018). Also, since these markers are distributed throughout the genome, it is feasible to identify those that correspond to genes subject to selection. Therefore, the movement of these variants through gene flow between individuals and populations is largely determined by the characteristics of the landscape (Creech et al. 2017). In this review we found that Combs et al. (2018) used thousands of markers (61,400 SNPs) to explore the patterns of spatial genetic structure of the Norway rat (Rattus norvegicus) in New York. The authors proved the existence of genetic structure at a fine spatial scale, attributed to the elements of the urban matrix and the ecological characteristics of the species. Also, these authors mention that previous studies, with the same species and using microsatellites, did not detect genetic structure patterns, highlighting the resolving power of SNPs in LG studies. On the other hand, Creech et al. (2017) explored the effects of landscape elements in gene flow from the optimization of resistance models, using simulations in populations of the bighorn sheep (a species that inhabits desert environments). Their results from simulations based on loci subject to selection show a higher gene flow of variants (loci) with adaptive potential in habitats with a continuous distribution of vegetation cover, among other variables. The above is particularly relevant in the context of the vulnerability of desert ecosystems (Bachelet et al. 2016; WWF 2019) and, therefore, the species that inhabit these systems, because their persistance will depend on the movement of genes with adaptive potential through an environment with scarce vegetation.
Remote sensing. One of the most powerful tools that has contributed to the generation of spatially explicit predictive variables is the information obtained from satellites, i. e., remote sensing (He et al. 2015). With our revision we evidence that one of the most innovative aspects in landscape genetics studies is the use of remote sensing information to achieve a more realistic interpretation of the landscape studied. For instance, Mapelli et al. (2012) used Landsat ETM+ sensor images to identify habitat patches in their study area and extract reflectance values of each, which were used to generate predictive variables. Thereby, the distance between good-quality patches was determined to be the most significant factor associated with the genetic structure patterns in Ctenomys porteousi. An additional sensor used to obtain satellite images is LiDAR, which has a higher resolution than Landsat, particularly for retrieving information on vegetation (Lefsky et al. 2002). Thus, Amaral et al. (2016) used LiDAR images to generate resistance layers used to assess the connectivity across populations of the rabbit Sylvilagus transitionalis and showed that patches of shrub vegetation facilitate gene flow in this species. As to its use in desert environments, Creech et al. (2014b) used MODIS (Moderate Resolution Imaging Spectroradiometer) images to extract reflectance values for vegetation and to estimate the NDVI, which is significantly associated with genetic diversity values in Ovis canadensis nelsoni; that is, vegetation determines the functional connectivity in this species. Flores-Manzanero et al. (2019), using Landsat 8 images, also calculated the NDVI for detecting the vegetation cover at a fine spatial scale, which produced various resistance models; the NDVI turned out to be the best predictor of gene flow for Dipodomys merriami. This strongly supports the conclusion that remote sensing is an excellent tool for landscape genetics studies, particularly in deserts, where the fine-scale definition of the various aspects of the landscape may be a complex issue.
Factors that determine genetic structure and diversity in mammals. The theoretical and methodological framework of population genetics is useful for inferring the patterns that govern the genetic structure and diversity of populations (Freeman and Herron 2002). Landscape genetics allows testing these inferences through hypothesis in a spatially explicit context (Manel et al. 2003; Holderegger and Wagner 2006; Balkenhol et al. 2015). The studies reviewed tested different environmental and landscape variables associated with genetic patterns in mammals. Geographic distance was the most significant variable and was best represented in rodents (California vole, Microtus californicus, Adams and Hadly 2010; pygmy rabbit, Brachylagus idahoensis, Thimmayya and Buskirk 2012; tuco tuco, Ctenomys lami, Lopes and De Freitas 2012; Córdoba vesper mouse, Calomys venustus, Chiappero et al. 2016), and in one carnivore (Panthera onca, Wultsch et al. 2016). Although in most studies anthropogenic impacts (fragmentation associated with agricultural land or roads) and low dispersal ability of species were identified as the drivers of the genetic patterns, we emphasize the importance of considering the greatest amount of environmental or ecological variables possible.
Also, most studies evaluated the effective distance from variables like topography (Vignieri 2005; Epps et al. 2007), vegetation cover (Munshi-South 2012;Lonsinger et al. 2015; Cullingham et al. 2016), precipitation and water bodies (Chambers and Garant 2010; Hapeman et al. 2011; Garrido-Garduño et al. 2016), available habitat, extent of fragmentation (Mapelli et al. 2012; Balkenhol et al. 2013), and anthropogenic impact (Gardner-Santana et al. 2009; Atwood et al. 2011; Amaral et al. 2016). Interestingly, some studies identify the ecological characteristics of species, in addition to environmental variables,as those that govern genetic diversity and structure patterns, particularly dispersal. For example, Chambers and Garant (2010) showed in the eastern chipmunk Tamias striatus that male-biased dispersal produces the structure pattern. Dispersal capability was also key in deserts, particularly for small mammals like Dipodomys spectabilis, since by incorporating population density data allowed detection of patterns that otherwise could have beeb attributed to geographic distance alone (Cosentino et al. 2015). In addition, one of the most significant factors for the bighorn sheep was maximum dispersal distance (i.e., effective distance), in addition to topography and food quality (estimated from the NDVI; Epps et al. 2007; Creech et al. 2014b). In fact, considering the maximum dispersal distance significantly improved the connectivity models, thus highlighting the importance of considering this little-used variable (Creech et al. 2014a). Finally, for large carnivores such as Ursus americanus, anthropogenic impact is the most important factor; for instance, the wall along the Mexico and USA border, is a significant barrier to dispersal and gene flow across populations (Atwood et al. 2011). Among medium-sized mammals like cacomixtle (Bassariscus astutus), the environment is the primary driver represented by the combination of vegetation type, slope, and elevation (Lonsinger et al. 2015).
North America… without Mexico? North America is the geographic region with the largest number of landscape genetics studies (Storfer et al. 2010). However, when Mexico was considered separately in our review, our analysis revealed the scarcity of studies despite its status as a megadiverse country that hosts a great variety of ecosystems (Mittermeier et al. 1997) and a high mammal richness, amounting to 496 species (Ramírez-Pulido et al. 2014). Although studies about genetics of the mammals of Mexico have been previously evaluated (Vázquez-Domínguez and Vega 2006), such assessment has not been done for landscape genetics. In this regard, it is important to mention that Rico (2019) recently published a review of landscape genetics studies conducted in Mexico to 2017, including 20 studies, with plants as the most studied taxonomic group (65%), while only two studies focused on mammals. This finding is consistent with our results, since we identified three studies for Mexico, two on rodents; of these, only one was conducted in a desert area. Garrido-Garduño et al. (2016) evaluated the effect of the landscape elements on the genetic structure and gene flow of Liomys pictus in a tropical deciduous forest, while Flores-Manzanero et al. (2019) determined the landscape features that influence gene flow of Dipodomys merriami in a region of the Chihuahuan desert. In Mexico, the tropical deciduous forest occupies 11.26 % of the national territory, mainly along the Pacific coast, while deserts represent 40% of the territory, distributed to the north and northwest (Challenger and Soberón 2008). Both ecosystems contain high levels of biodiversity and endemisms, particularly of animals (CONANP 2006; Ceballos et al. 2010); this, together with spatial and temporal heterogeneity, makes them suitable systems for hypothesis testing in a landscape genetics context.
Prospects for landscape genetics studies: the case of Mexico. From our review outlined here, we can assert that it is imperative in Mexico to conduct research addressing the effects of landscape and the environment on the distribution of genetic variation and structure of wild populations, through a spatially explicit approach. However, since rodents were the order of mammals most represented in this review, highlighting the studies conducted in Mexico, we believe landscape genetics studies with rodents will likely increase in the near future. This is based on the diversity of studies conducted with rodents in different environments in Mexico focused on aspects of genetic structure and diversity (Vega et al. 2007; Castañeda-Rico et al. 2011; Espindola et al. 2014), taxonomy and systematics (Arellano et al. 2006; Álvarez-Castañeda et al. 2009; Fernández et al. 2012), phylogeography (Espinoza-Medinilla et al. 2006; Gutiérrez-García and Vázquez-Domínguez 2012; Álvarez-Castañeda and Murphy 2014), diversification and speciation (Castañeda-Rico et al. 2013; Pérez-Consuegra and Vázquez-Domínguez 2015), and even the development of molecular markers (Munguía-Vega et al. 2007; Vázquez-Domínguez and Espindola 2013), to mention a few. In addition, the development of environmental and climatic layers with better resolution for Mexico (Téllez-Valdés et al. 2010; Cuervo-Robayo et al. 2014) will be key for the development of these studies in the country, in combination with remote sensing data that are currently freely accessible (Landsat 8; http://landsat.usgs.gov/). Finally, deserts cover a large part of the national territory (ca. 70 million hectares; Challenger and Soberón 2008) and in some cases, as in the Chihuahuan desert, vegetation is patchily distributed (Grünberger 2004). It is worth highlighting that, although deserts can be thought of as relatively homogeneous systems, in reality the spatial distribution of the elements of the desert landscape renders a heterogeneous matrix that allows evaluating the structural and functional connectivity (Manel and Holderegger 2013), making deserts ideal ecosystems to conduct landscape genetics studies.
Some of the most significant advances for the study of landscape genetics in Mexico and elsewhere are worth mentioning, including the use of remote sensing data and species distribution models, which yield a better representation of the landscape and help to set hypotheses considering structure and connectivity within a spatially explicit context. The use of genomic tools (markers, bioinformatics methods, and analyses) and adaptive approaches will allow addressing questions not only regarding the effect of the landscape on genetic patterns but also about how individuals respond in terms of adaptation and selection.











 text new page (beta)
text new page (beta)


