Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias pecuarias
On-line version ISSN 2448-6698Print version ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.14 n.1 Mérida Jan./Mar. 2023 Epub Mar 24, 2023
https://doi.org/10.22319/rmcp.v14i1.6121
Articles
Changes in the count of four bacterial groups during the ripening of Prensa (Costeño) Cheese from Cuajinicuilapa, Mexico
aUniversidad Autónoma Chapingo. Departamento de Ingeniería Agroindustrial. Carretera México-Texcoco km 38.5, Texcoco, Estado de México. México.
bUniversidad de Guadalajara. Centro Universitario de Ciencias Biológicas y Agropecuarias, Zapopan, Jal. México.
c Universidad Autónoma Chapingo. Departamento de Preparatoria Agrícola. Texcoco, Estado de México. México.
Prensa Cheese, also called Costeño, is made in an artisanal way from raw cow’s milk in the Costa Chica region of the state of Guerrero. In order to know the characteristics of Mexican artisanal cheeses, the objective of this research was to analyze the changes in the count of aerobic mesophilic bacteria (AMB), total coliform (TCs) microorganisms, lactic acid bacteria (LAB) and coagulase-positive staphylococci (CPS), during the ripening (5, 30, 60 and 90 d) of Prensa cheeses, made by four different cheese factories (A, B, C and D) of Cuajinicuilapa, Guerrero, Mexico. A portion (25 g) of each cheese sample was homogenized with peptone diluent (225 mL) and dilutions from 10-1 to 10-6 were prepared with which 3MTM PetrifilmTM plates were sown. After incubating under different conditions, depending on the type of microorganism, AMB, TCs, LAB and CPS counts were made. The results showed that as the ripening time of the Prensa Cheese progressed, the microbial load decreased: AMB from 4 to 2, TCs from 6 to 3, LAB from 6 to 2 and CPS from 5 to 2 log10 CFU g-1. The changes in the counts of the bacterial groups studied can be attributed to the physicochemical and microbiological transitions typical of cheese maturation and to the characteristics of the microbiota present in each of the cheese factories. The results of this research provide elements for the microbial characterization of Mexican artisanal cheeses.
Key words Lactic acid bacteria; Aerobic mesophilic bacteria; Coagulase-positive staphylococci; Raw milk; Microbiota; Total coliform microorganisms; Artisanal cheeses
El Queso de Prensa, también llamado Costeño es elaborado artesanalmente a partir de leche cruda de vaca en la región de la Costa Chica del estado de Guerrero. Con el fin de conocer las características de los quesos artesanales mexicanos, el objetivo de esta investigación fue analizar los cambios en el recuento de bacterias mesófilas aerobias (BMA), microorganismos coliformes totales (CT), bacterias ácido lácticas (BAL) y estafilococos coagulasa positivos (ECP), durante la maduración (5, 30, 60 y 90 días) de quesos de prensa, elaborados por cuatro diferentes queserías (A, B, C y D) de Cuajinicuilapa, Guerrero, México. Una porción (25 g) de cada muestra de queso se homogeneizó con diluyente de peptona (225 ml) y se prepararon diluciones desde 10 -1 a 10 -6 con las que se sembraron placas 3M TM PetrifilmTM. Después de incubar a diferentes condiciones, según el tipo de microorganismo, se hicieron recuentos de BMA, CT, BAL y ECP. Los resultados mostraron que conforme avanzó el tiempo de maduración del Queso de Prensa, la carga microbiana disminuyó: BMA de 4 a 2, CT de 6 a 3, BAL de 6 a 2 y ECP de 5 a 2 log10 UFC g-1. Los cambios en los recuentos de los grupos bacterianos estudiados, pueden ser atribuidos a las transiciones fisicoquímicas y microbiológicas propias de la maduración del queso y a las características de la microbiota presente en cada una de las queserías. Los resultados de esta investigación aportan elementos para la caracterización microbiana de los quesos artesanales mexicanos.
Palabras clave Bacterias ácido lácticas; Bacterias mesófilas aerobias; Estafilococos coagulasa positivos; Leche cruda; Microbiota; Microorganismos coliformes totales; Quesos artesanales
Introduction
Around the world, cheese, in addition to being a rich source of nutrients, is an essential food used in the local gastronomy of different societies1,2. Currently, around 1,833 varieties of cheese located in 74 countries are known3; this diversity is determined by the technological processes used for its preparation, such as the origin of milk, fat-protein ratio, types of cultures and coagulating agents; the shape, size of the cheese and the maturation conditions4,5,6.
Cheese ripening consists of its storage, under certain conditions of temperature and moisture, for a period of time that can range from 3 to 7 d, up to 2 yr5,7. The maturation process, in addition to providing sensory characteristics, is a method of conservation5,6,8. In this stage, biotic and abiotic changes that have a direct impact on the microbiota present in the cheese occur5,7,9.
Most artisanal cheeses are made from raw milk, with spontaneous fermentation, non-technified preparation processes and varied maturation times5,7,10.
In Mexico there are about 40 artisanal cheeses7, among them are matured cheeses such as Cotija from the Sierra JalMich, Añejo cheese from Zacazonapan, Maduro cheese from Veracruz, Chihuahua cheese and artisanal cheese from the Ojos Negros region of Baja California, of which some aspects of their microbiology have been published11-14. Prensa cheese (PC) is made with unpasteurized cow’s milk, commercial liquid rennet and salt; it goes through a pressing stage whose duration varies at the discretion of the manufacturer from 1 to 3 days, then it is left to mature for periods of up to three months. The cheese thus obtained has color variations between white and yellow15. It is generally rectangular or circular in shape, its consistency is firm, and its weight is 1 to 14 kg per piece (Figure 1)15.
PC has been produced for more than 100 years in southwestern Mexico, mainly in the Costa Chica region of the state of Guerrero in the municipalities of Cuajinicuilapa and Ometepec, as well as in the municipality of Pinotepa Nacional, on the coast of the state of Oaxaca15. According to INEGI16, the climate of the Costa Chica region is warm subhumid, and its temperatures range from 22 to 28 °C.
Studies are currently being carried out to identify the characteristics of artisanal cheeses from Mexico7,15, the objective of this research was to analyze the changes that occur in the count of aerobic mesophilic bacteria, total coliform microorganisms, lactic acid bacteria and coagulase-positive staphylococci, during the ripening (5, 30, 60 and 90 d) of prensa cheeses made by four cheese factories (A, B, C and D) of Cuajinicuilapa, Guerrero, Mexico.
Material and methods
Cheese samples
Samples of PC made in an artisanal way in the municipality of Cuajinicuilapa, Guerrero, Mexico (16°28′18′ N, 99°24′55′ W), were analyzed in July 2018. Based on a targeted sampling, four cheese factories were selected as sampling units, which will henceforth be named A, B, C and D. Four freshly made cheeses, weighing 1 kg, were purchased from each cheese factory. The samples were moved to the municipality of San Marcos, Guerrero (16°47′46′ N, 99°23′05′ W) to a space with characteristics similar to those of the cheese factories of Cuajinicuilapa. In this place, the samples of the cheeses from the cheese factories A, B, C and D (four of each cheese factory) were left to mature for 5, 30, 60 and 90 d. After the ripening time, each batch was transferred in polyethylene bags, inside coolers with refrigerant, to the laboratory. The samples were kept in refrigeration at 4 °C until analysis. The maximum ripening time was 90 d because after that period the flavors intensify, and local consumers avoid it because they prefer softer flavors.
Sample preparation
Each of the cheese samples (25 g) was mixed with 225 mL of peptone diluent, the mixture was homogenized for 2 min (VWR® symphony D S41 Vortex, VWR International) and dilutions from 10-1 to 10-6 were made by transferring 1 mL of the sample to vials containing 9 mL of peptone diluent17.
Microorganism count
The following culture media (3M PetrifilmTM plates) were used: aerobe count (AC No. of catalog 6400), coliform count (TC No. of catalog 6410), lactic acid bacteria (No. of catalog 6461) and staph express for coagulase-positive staphylococci (No. of catalog 6493); 1 mL of the corresponding dilution was placed in each of the plates18-21.
All counts were done in duplicate. For AMB, dilutions 10-3 and 10-4 were sown and the medium was incubated at 35 ± 2 °C for 48 ± 3 h18. TC microorganisms were studied based on dilutions 10-2 and 10-3, being incubated at 35 ± 1 °C for 24 ± 2 h19. The determination of LAB was made by inoculating the media with dilutions from 10-3 to 10-6 and incubating at 35 ± 2 °C for 48 ± 3 h20. The CPS study was conducted from dilutions 10-2 to 10-4 and an incubation at 37 ± 1 °C for 24 ± 3 h21.
Once the incubation time was completed, the growth was reviewed and the plates containing between 15 and 300 colonies were counted, the mean of the two repetitions was obtained and this average was multiplied by the inverse of the dilution with which the plate was inoculated22. The result of the count was reported as log10 of the number of colony-forming units per gram (log10 CFU g-1)23.
Statistical analysis
The statistical analysis was based on a design with repeated means and completely random distribution of treatments, tested over time, whose probabilistic model corresponds to:
Where:
The effect of the treatments (cheese factories A, B, C and D) was evaluated through the ripening time (5, 30, 60 and 90 d) with four repetitions, generating 64 experimental units, each consisting of 25 g of cheese.
The response variables evaluated were: total count of aerobic mesophilic bacteria (AMB), total coliforms (TCs), lactic acid bacteria (LAB) and coagulase-positive staphylococci (CPS). The data were analyzed using a mixed model24,25 whose random effect corresponds to the maturation time and the fixed effect to the cheese factories. The Tukey-Kramer method (P<0.05) was applied to identify the effect of the treatments. Analyses were performed in the SAS package version 9.1 (SAS Institute, Inc., Cary, NC, USA).
Results and discussion
During the time of ripening, a decrease in the count of the different microorganisms was observed. The highest counts of all microbial groups were reached at 5 d of ripenig while the minimum values occurred at 90 d (Table 1).
Table 1 Count of bacterial groups, log10 CFU g-1, during the ripening of Prensa cheese made in four cheese factories (A, B, C and D) of Cuajinicuilapa, Guerrero, Mexico
| Bacterial group | Time (days) | Cheese factories | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Aerobic mesophilic bacteria (log10 CFU g-1) | 5 | 6.3143 ±0.2973 Cc | 4.6802 ±0. 2973 Ba | 5.8948 ±0.2973 Cb | 5.9077 ±0.2973 Cb |
| 30 | 5.1156 ±0.2973 Bb | 4.3884 ±0. 2973 Ba | 4.4981 ±0.2973 Ba | 5.1043 ±0.2973 Bb | |
| 60 | 4.0021 ±0.2973 Ab | 3.1505 ±0. 2973 Aa | 4.4047±0.2973 Bbc | 4.6703±0.2973 ABc | |
| 90 | 4.3178 ±0.2973 Ab | 2.5638 ±0. 2973 Aa | 2.4005 ±0.2973 Aa | 4.3597 ±0.2973 Ab | |
| Total coliforms (log10 CFU g-1) | 5 | 4.1093 ±0.1002 Cb | 2.1505 ±0.1002 Aa | 2.4203 ±0.1002 Aa | 4.7211 ±0.1002 Db |
| 30 | 3.7726 ±0.0541 Cc | 2.8838 ±0.0541 Ba | 3.7916 ±0.0541 Cc | 3.0878 ±0.0541 Cb | |
| 60 | 2.3138 ±0.2854 Bb | 2.3763 ±0.2854 ABb | 2.2698 ±0.2854 Ab | 1.5753 ±0.2854 Ba | |
| 90 | <1 ±0.0440 Aa | 2.3451 ±0.0440 Ab | 2.0753 ±0.0440 Ab | <1 ±0.0440 Aa | |
| Lactic acid bacteria (log10 CFU g-1) | 5 | 6.5457 ±0.0651 Dc | 5.9454 ±0.0651 Cb | 5.5084 ±0.0651 Ba | 6.7366 ±0.0651 Cc |
| 30 | 5.8336 ±0.0651 Cb | 5.8231 ±0.0651 Cb | 5.1193 ±0.0651 Ba | 6.1241 ±0.0651 Cb | |
| 60 | 3.5524 ±0.0651 Ba | 3.7918 ±0.0651 Ba | 3.1945 ±0.0651 Aa | 5.0914 ±0.0651 Bb | |
| 90 | <1 ±0.0651 Aa | <1 ±0.0651 Aa | 3.2258 ±0.0651 Ab | 4.2394 ±0.0651 Ac | |
| Coagulase-positive staphylococci (log10 CFU g-1) | 5 | 5.6562 ±0.0540 Cb | 3.7456 ±0.0540 Ba | 5.6918 ±0.0540 Cb | 5.8746 ±0.0540 Cb |
| 30 | 3.6276 ±0.3811 Bb | 2.2500 ±0.3811 Aa | 3.7271 ±0.3811 Bb | 3.8389 ±0.3811 Bb | |
| 60 | 2.6945 ±0.0438 Aa | 2.7143 ±0.0438 Aa | 2.7311 ±0.0438 Aa | 2.8063 ±0.0438 Aa | |
| 90 | 2.5951 ±0.0524 Aa | 2.4203 ±0.0524 Aa | 2.6945 ±0.0524 Aa | 2.5951 ±0.0524 Aa | |
Means with lowercase letter in rows and means with uppercase letter in columns, followed by different letter, indicate statistical significance (Tukey, P<0.05).
Aerobic mesophilic bacteria
In the AMB counts, there were significant differences (P<0.05) between the cheese factories (Table 1). The gradual decrease in AMB from day 5 to 90 was close to 2 log10 CFU g-1, for cheese factories A, B and D; while for cheese factory C, it was 3.49 log10 CFU g-1.
Most of the AMB values found in the PC are within the range of 4 to 9 logarithms CFU g-1, reported for cheeses made from raw milk and matured for 60 or more days26. Since the maturation process of cheeses involves the multiplication of the microorganisms present, AMB concentrations of 4 to 9 logarithms (104 to 109 CFU g-1) are expected in this type of products, without this implying a deterioration of the food or suggesting that non-sanitary conditions occurred during its preparation or storage26,27,28. In the region of Ojos Negros in the state of Baja California, based on a study that included matured cheeses from 22 cheese factories, made with raw milk, it is reported that AMB were found in a range of 4.6 to 7.2 log10 CFU g-1(14.
In studies on the ripening of artisanal Cotija cheese, salted and matured at temperatures of 14 °C to 32 °C, Chombo11 reports the following variations in AMB: 8.3, 7.0, 3.5 and 4.7 log10 CFU g-1, on d 8, 30, 60 and 90, while Magallón29 found 5.3 and 1.8 log10 on d 30 and 90, respectively. The counts found in the PC at 30 and 90 d are very close to those reported for Cotija cheese that is ripened in temperature ranges similar to those of PC.
The decrease in AMB was common in the cheeses from the four cheese factories (Table 1), reflecting a certain homogeneity in the preparation processes and the ripening conditions of the cheeses. The statistical difference (P<0.05) between cheese factories suggests quantitative or qualitative variations in the microbiota of cheese generated by milk and the microenvironments of each cheese factory. It should be noted that, between d 5 and 60, it is observed that the AMB of cheeses from cheese factory A descend 2.31 log10 CFU g-1, however, between d 60 and 90 they remain unchanged; while the AMB of cheeses from cheese factory C, from d 60 to 90, show a reduction of 2 log10 CFU g-1.
The development of AMB in cheeses from cheese factory A shows the ability of bacteria to adapt and survive, while the decrease in AMB in the cheeses from cheese factory C exhibits a loss of viability with the release of enzymes that contribute to the generation of flavors and textures5. This suggests that it is appropriate to study the relationship of the organoleptic characteristics of prensa cheese, between cheese factories and between different concentrations of AMB over maturation time, as well as the relationship of AMB with the shelf life of cheese at room temperature. On the other hand, AMB studies are useful as an initial stage in the search for starter cultures from artisanal cheeses. Variations in salt concentration and moisture may have influenced the survival of AMB. The results suggest that, in cheeses from cheese factories A and D, there are bacteria adapted for long-term survival. While in cheeses from cheese factories B and C, bacteria of short survival may be present, and therefore can contribute more quickly to the production of tastes, smells and textures (pleasant or unpleasant)5. Another possibility is that the reduction of AMB in cheeses from cheese factories B and C is due to the presence of substances with antimicrobial action, caused by the metabolism of microorganisms, or by the biochemical changes that occur in the cheese, derived from the proteolysis of casein to give rise to peptides with antimicrobial activity5,30.
Total coliform bacteria
Total coliform counts in the cheeses showed significant differences (P<0.05) between cheese factories during the ripening period (Table 1). The highest counts were found on d 5 (4.72 log10 CFU g-1) and the lowest on d 90 (<1 log10). Cheese, being a solid sample, hinders a direct count, so it is necessary to make an initial dilution that leads to the minimum detection level being 10 CFU g-1, so the absence of growth was reported as < 1 log10.
Two different dynamics were observed, cheeses from cheese factories A and D had the highest initial TC loads, which decreased at 30 d of maturation to reach 3.72 to 3.10 log10 CFU g-1, respectively (Table 1). In cheeses from cheese factories B and C, the TCs showed initial levels of 2 logarithms, which rose during the first month and remained with slight variations to coincide on d 90 with values very close to each other.
The dynamics of TCs in cheeses from cheese factories A and D have been reported in semi-hard cheeses and are characterized by a progressive decrease in coliforms as ripened progresses31, which is attributed to the decrease in pH due to the fermentation of lactose32. In ripened cheeses such as Cheddar, coliforms die at a rate of 0.3 log10 CFU g-1 per week and in Gouda cheese at 0.7 log10 per week33. Therefore, the dynamics observed in cheeses from cheese factories B and C is atypical, because between d 5 to 30, there is an increase of 0.73 and 1.37 log10, respectively, followed on d 60 to 90 by very small decreases, 0.03 and 0.19 log10, respectively (Table 1). This suggests that there are common aspects between the two cheese factories that favor the selection of bacteria that persist during ripening, such as water and milk quality, personnel or variations in the preparation or cleaning processes.
The accepted levels of total coliforms in matured cheeses are less than 100 CFU/g (< 2 log10 CFU g-1)34, values that were reached between d 60 and 90 (Table 1) in cheese factories A and D. According to Metz32, cheeses made with good quality raw milk, under hygienic sanitary conditions, applying good manufacturing practices and properly ripened, will have low levels of total coliform bacteria, fecal coliforms, enterococci, Enterobacteriaceae and Escherichia coli.
The activity of coliform organisms in cheeses can adversely affect their sensory characteristics28,35. However, it has been observed that certain genera of coliforms contribute positively to the texture and sensory characteristics of the product; in addition to the fact that some strains of Hafnia contribute to the accumulation of aromas, and generation of flavors32,35,36. The persistence of coliforms suggests the possibility of the participation of these microorganisms in the organoleptic characteristics of cheeses from cheese factories B and C, an aspect that has not been addressed in studies of Mexican cheeses.
Lactic acid bacteria
During the ripening period, the LAB counts of the cheeses from the cheese factories studied showed significant differences (P<0.05) (Table 1). This microbial group had the highest counts of the entire study. On d 5 the counts ranged between 5.50 and 6.73 log10 CFU g-1, these values decreased from day 30. From day 5 to 60, the reductions were 2.99, 2.15, 2.31 and 1.77 log10, for cheeses from cheese factories A, B, C and D, respectively.
During the ripening of the Spanish artisanal cheeses Casar de Cáceres, Afuega’l Pitu and Cabrales, decreases in lactococcal counts of 2 to 3 log10 CFU g-1 were reported between d 0 and 60, while in “La Serena” cheese there was only a reduction of less than one logarithm, this is attributed to the fact that this cheese had a low salt content during the first weeks of ripening5. This suggests that the reductions observed in the PC from d 5 to 60 are consistent with what happens with homofermentative lactic acid bacteria in cheeses made in an artisanal way with native microbiota5. The concentrations of LAB found in the PC from d 60 to 90 are lower than those reported in ripened cheeses from Europe, which have values of 7 to 9 log10 CFU g-1(5,37,38.
With differences in the type of cattle and geographical areas, Cotija cheese and PC share temperatures, preparation and ripening processes. In Cotija cheese, small increases and decreases have been found in LAB counts; one study reports 2.6 log10 CFU g-1 on d 30 with an increase to 2.9 log10 on d 9029, another study indicates 5.9 log10 on d 60, which decreases to 5.0 log10 on d 9011. The above data suggest that, in both PC and Cotija cheese, LAB counts tend to be lower than in other ripened cheeses; this could be explained by the temperatures at which they are ripened, which favors a greater loss of moisture that generates values of water activity and moisture/salt ratio that are inhibitory for LAB5.
Coagulase-positive staphylococci
The CPS counts of the cheeses from the cheese factories studied showed significant differences (P<0.05) during the ripening period (Table 1). From d 5 to 60, a continuous decrease in CPS was observed in cheeses from cheese factories A, C and D, followed by a stabilization from d 60 to 90. In cheese factory B, there was a decrease from d 5 to 30 followed by an increase from d 30 to 60 and a stabilization from d 60 to 90 (Figure 2). The death rates of CPS (average decrease log10 CFU g-1 divided between week of ripening)39 in cheeses from cheese factories A and C were 0.30 log10 and 0.23 and 0.31 log10 in cheese factories B and D, respectively.
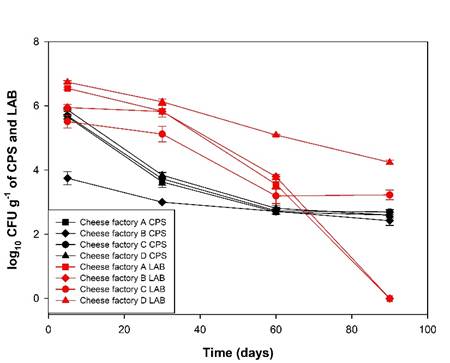
Figure 2 Antagonism and change in lactic acid bacteria (LAB) concentration with respect to coagulase-positive staphylococci (CPS)
For cheeses made from raw milk, the accepted limits of coagulase-positive staphylococci are 104 to 105 CFU g-1, which is equivalent to 4-5 log10 CFU g-140, limits that were exceeded on day 5 in cheese factories A, C and D but that were reached again on day 30 and remained until day 90 (Table 1). CPS counts greater than 4 log10 show the need to apply corrective measures in the hygiene of the processes of milk collection, cheese making and the selection of raw materials40. Values of 105 CFU g-1 or higher lead to study the presence of staphylococcal toxin in cheeses40, since being thermostable, it can persist even when staphylococci have died. Concentrations of 106 CFUs g-1 are usually needed to produce enough toxin (one nanogram per gram of cheese) to cause a disease outbreak5.
Reductions of Staphyloccocus aureus of 1 to 3 logarithms have been reported in different cheeses41,42,43, figures that coincide with reductions in CPS during PC ripening.
The death rate of S. aureus in Manchego cheese made with raw milk from d 1 to 60 is 0.404 log10 CFU g-139, the death rates of CPS in cheese factories A, C and D (0.49, 0.50 and 0.52) were close to this value, suggesting that the decrease was as expected for this type of cheese (matured Prensa paste cheeses). No factors that explain the lower death rate observed in cheese factory B (0.36) were identified.
The development and survival of S. aureus are affected by factors such as: physicochemical changes that occurred during the ripening process, secondary metabolites generated by LAB, as well as the composition of the product, storage period and temperature44,45. Staphyloccocus aureus is inhibited by LAB through nutrient competition, production of lactic acid, hydrogen peroxide and production of antimicrobial substances46, which may explain the decrease in CPS in the first 30 d of ripening, a period in which LAB levels were higher (Figure 2). Between d 30 and 60, storage conditions at room temperature could increase moisture loss, which changes the moisture/salt ratio, being inhibitory for LAB5, this favors S. aureus, which could explain its slight increase in cheese factory B and the suspension of its decrease in cheese factories A, C and D.
Conclusions and implications
Prensa cheese is a cheese made in an artisanal way, ripened in warm subhumid climate with the participation of native lactic acid bacteria, whose concentrations are lower than those reported for European cheeses, but similar to those reported in Cotija cheese. Statistical differences in microbial counts at different times show the changes that occur as cheese matures. Meanwhile, the statistical differences between the cheese factories suggest the existence of microbiomes specific to each cheese factory, which could be able to generate variants of PC among different artisanal producers, even when they have similar production processes. The changes in the counts of the bacterial groups studied can be attributed to physicochemical changes and successions in the bacterial populations typical of the maturation of the cheese and to the characteristics of the microbiota present in each of the cheese factories. It is convenient to explore whether the shelf life of this cheese extends beyond 90 days. The finding of coliforms that persist during ripening shows the need to investigate whether this is an exceptional case, and bacteria from this group contribute to the pleasant characteristics of the cheese or are related to its deterioration. The data on reduction and survival of coagulase-positive staphylococci generated in this research can serve as a reference to initiate and evaluate improvement programs in this type of cheese factories. Although this research included four cheese factories in the main PC-producing municipality, the information generated can serve as a reference for the characterization of this artisanal cheese.
Acknowledgements and conflicts of interest
This research work was possible thanks to the scholarship granted by the National Council of Science and Technology for the master’s studies of the first author. We also appreciate the comments made on the manuscript by Doctors Angélica Luis Juan Morales and Ricardo Alaniz de la O. None of the authors has any conflict of interest with respect to this publication.
REFERENCES
1. Cidón CD, Canut E. Quesos españoles, ases en la mesa, comodines en la cocina. 1a ed. Madrid, ESP: Everest; 2003. [ Links ]
2. Yescas C, Santacruz J. Quesos mexicanos. 1a ed. México, Distrito Federal: Larousse; 2013. [ Links ]
3. Cheese.com specialty cheeses. Alphabetical list, find over 1833 specialty cheeses from 74 countries in the world´s greatest cheese resource. https://www.cheese.com/alphabetical/ . Accessed Jan 26, 2021. [ Links ]
4. Kongo JM, Malcata FX. Cheese: Types of cheeses - soft. In: Caballero B, Toldrá F, Pinglas PM, editors. Encyclopedia of food and health. 1rst ed. Oxford, GB: Academic Press; 2015:768-773. [ Links ]
5. Fox PF, Guinee TP, Cogan TM, McSweeney PLH. Fundamentals of cheese science. 2nd ed. New York, USA: Springer; 2017. [ Links ]
6. Santiago-López L, Aguilar-Toalá JE, Hernández-Mendoza A, Vallejo-Cordoba B, Liceaga AM, González-Córdova AF. Invited review: Bioactive compunds produced during cheese ripening and health effects associated with aged cheese consumption. J Dairy Sci 2018;101(5):3742-3757. [ Links ]
7. Villegas GA, Santos MA, Cervantes EF. Los quesos mexicanos tradicionales. 1a ed. Texcoco, MEX: Universidad Autónoma Chapingo; 2016. [ Links ]
8. Alejo-Martínez K, Ortiz-Hernández M, Recino-Metelin BR, González-Cortés N, Jiménez-Vera R. Tiempo de maduración y perfil microbiológico del queso de poro artesanal. ReIbCi 2015;2(5):15-24. [ Links ]
9. Sánchez-Valdés JJ, Colín-Navarro V, López-González F, Avilés-Nova F, Castelán-Ortega OA, Estrada-Flores JG. Diagnóstico de la calidad sanitaria en las queserías artesanales del municipio de Zacazonapan, Estado de México. Salud Publ Mex 2016;58(4):461-467. [ Links ]
10. Vasek O, Cardozo M, Fusco AJ. Producción artesanal de quesos. Sistema de transformación agroalimentario en la región correntina (Argentina). IV Congreso internacional de la red SIAL. Mar del Plata, provincia de Buenos Aires.2006:1-32. [ Links ]
11. Chombo-Morales P, Kirchmayr M, Gschaedler A, Lugo-Cervantes E, Villanueva-Rodríguez S. Effects of controlling ripening conditions on the dynamics of the native microbial population of Mexican artisanal Cotija cheese assessed by PCR-DGGE. LWT-Food Sci Technol 2016;65:1153-1161. [ Links ]
12. Ramírez-Rivera EJ, Ramón-Canul LG, Torres-Hernández G, Herrera-Corredor JA, Juárez-Barrientos JM, Rodríguez-Miranda J, et al. Tipificación de quesos madurados de cabra producidos en la zona montañosa central del estado de Veracruz, México. Agrociencia 2018;52(1):15-34. [ Links ]
13. Sánchez-Gamboa C, Hicks-Pérez L, Gutiérrez-Méndez N, Heredia N, García S, Nevárez-Moorillón GV. Microbiological changes during ripening of Chihuahua cheese manufactured with raw milk and its seasonal variations. Foods 2018; 7(9):153. https:// www.mdpi.com/2304-8158/7/9/153 . Accessed Jan 30, 2021. [ Links ]
14. Silva-Paz LE, Medina-Basulto G. E., López-Valencia G, Montaño-Gómez MF, Villa-Angulo R, et al. Caracterización de la leche y queso artesanal de la región de Ojos Negros, Baja California, México. Rev Mex Cienc Pecu 2020;11(2):553-564. https://cienciaspecuarias.inifap.gob.mx/index.php/Pecuarias/article/view/5084 . Consultado 30 Ene, 2021. [ Links ]
15. Sandoval-Alarcón F. Caracterización y análisis de la productividad del queso de prensa de la Costa Chica de Guerrero y Oaxaca [tesis maestría]. Texcoco, Estado de México: Universidad Autónoma Chapingo; 2016. [ Links ]
16. INEGI. Instituto Nacional de Estadística y Geografía. Guerrero, Clima. Recuperado de: Recuperado de:http://www.cuentame.inegi.org.mx/monografias/informacion/gro/territorio/clima.aspx?tema=me&e=12 . Consultado 02 Sep, 2020. [ Links ]
17. Romero-Castillo PA, Leyva-Ruelas G, Cruz-Castillo JG, Santos-Moreno A. Evaluación de la calidad sanitaria de quesos crema tropical mexicano de la región de Tonalá, Chiapas. Rev Mex Ing Quím 2009;8(1):111-119. [ Links ]
18. 3M México. Placas Petrifilm™ para el recuento de aerobios AC, guía de interpretación. Ciudad de México, México: 3M. 2017. [ Links ]
19. 3M México. Placas Petrifilm™ para el recuento coliformes, guía de interpretación. Ciudad de México, México: 3M. 2017. [ Links ]
20. 3M México. Placas para el recuento de bacterias ácido lácticas 3M® Petrifilm®, guía de interpretación. Ciudad de México, México: 3M. 2017. [ Links ]
21. 3M México. Placas Petrifilm™ Staph Express para recuento de Staphylococcus aureus, guía de interpretación. México DF, México: 3M. 2009. [ Links ]
22. Camacho A, Giles M, Ortegón A, Palao M, Serrano B, Velázquez O. Técnicas para el análisis microbiológico de alimentos, cuenta en placa de bacterias. 2a ed. México, DF, México: Facultad de Química.; 2009. http://depa.fquim.unam.mx/amyd/archivero/TecnicBasicas-Cuenta-en-placa_6527.pdf . Consultado 2 Sep, 2020. [ Links ]
23. FAO, WHO. Statistical aspects of microbiological criteria related to foods, a risk managers guide. 1st ed. Rome, ITA: Food and Agriculture Organization of the United Nations; 2016. [ Links ]
24. Little RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. SAS® for mixed models. 2nd ed. Cary, North Carolina, USA: SAS Institute Inc; 2006. [ Links ]
25. Stroup WW, Milliken GA, Claassen EA, Wolfinger RD. SAS® for mixed models: Introduction and basic applications. 3rd ed. Cary, North Carolina, USA: SAS Institute Inc ; 2018. [ Links ]
26. Brooks JC, Martinez B, Stratton J, Bianchini A, Kroksrom R, Hutkins R. Survey of raw milk cheeses for microbiological quality and prevalence of foodborne pathogens. Food Microbiol 2012;31(2):154-158. [ Links ]
27. ICMSF. International Commission on Microbiological Specifications for Foods. Microorganismos de los alimentos 1. Técnicas de análisis microbiológico. 2da ed. Zaragoza, ESP: Acribia; 2000. [ Links ]
28. Fernández-Escartín E. Microbiología e inocuidad de los alimentos. 2da ed. Querétaro, MEX: Universidad Autónoma de Querétaro; 2008. [ Links ]
29. Flores-Magallón R, Oliva-Hernández AA, Narváez-Zapata AA. Characterization of microbial traits involved with the elaboration of the Cotija cheese. Food Sci Biotechnol 2011;20(4):997-1003. [ Links ]
30. López-Expósito I, Miralles B, Amigo L, Hernández-Ledesma B. Health effects of cheese components with a focus on bioactive peptides. In: Frias J, Martinez-Villaluenga C, Peñas E, editors. Fermented foods in health and disease prevention. 1st ed. London, UK: Academy Press; 2017:239-273. [ Links ]
31. Asperger H, Brandl E. The significance of coliforms as indicator organisms in various types of cheese. Antonie van Leeuwenhoek 1983;48:635-639. [ Links ]
32. Metz M, Sheehan J, Feng PCH. Use of indicator bacteria for monitoring sanitary quality of raw milk cheeses - A literature review. Food Microbiol 2020;85(2020): 103283. https://www.sciencedirect.com/science/article/abs/pii/S0740002018311213?via%3Dihub . Accesed Jan 30, 2021. [ Links ]
33. Fox PF, Guinee TP, Cogan TM, McSweeney PLH. Fundamentals of cheeses science. 1st ed. Gaithersburg, Maryland, USA: Aspen Publisher 2000. [ Links ]
34. NACMCF. National Advisory Committe on Microbiological Criteria for Foods. NACMCF-Report-Process-Control-061015 (1) response to questions posed by the Department of Defense regarding microbiological criteria as indicators of process control or insanitary conditions, Washington DC, USA: United States Department of Agriculture; 2015. https://www.fsis.usda.gov/sites/default/files/media_file/2020-07/NACMCF-Report-Process-Control-061015.pdf . Accesed Aug 07, 2021. [ Links ]
35. Martin NH, Trmčić A, Hsieh TH, Boor KJ, Wiedmann, M. The evolving role of coliforms as indicators of unhygienic processing conditions in dairy foods. Front Microbiol 2016;7:1549. https://www.frontiersin.org/articles/10.3389/fmicb.2016.01549/full . Accessed Aug 07, 2021. [ Links ]
36. Trmčić A, Chauhan K, Kent DJ, Ralyea RD, Martin NH, Boor KJ, et al. Coliform detection in cheese is associated with specific cheese characteristics but no association was found with pathogen detection. J Dairy Sci 2016;99(8):6105-6120. [ Links ]
37. Khalid NM, Marth EH. Lactobacili - their enzymes and role in ripening and spoil age of cheese- A review. J Dairy Sci 1990;73(10):2669-2684. [ Links ]
38. Tavaria FK, Reis PJM, Malcata FX. Effect of dairy farm and milk refrigeration on microbiological and microstructural characteristics of matured Serra da Estrella cheese. Int Dairy J 2006;16(8):895-902. [ Links ]
39. Nuñez M, Bautista L, Medina M, Gaya P. Staphylococcus aureus, thermostable nuclease and staphylococcal enterotoxins in raw ewes' milk Manchego cheese. J Appl Bacteriol 1988;65(1):29-34. [ Links ]
40. Commission Regulation (EC) No. 2073/2005 of 15 November 2005 on microbiological criteria of foodstuffs L338. Official Journal of the European Union 2005 L338: 1-26. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32005R2073 Accessed Sep 2, 2020. [ Links ]
41. Kirdar SS, Yurdakul O, Kalit S, Kalit M. Microbiological changes throughout ripening of Keş cheese. J Cent Eur Agric 2018;19(1):61-71. [ Links ]
42. Cardoso VM, Dias RS, Soares BM, Clementino LA, Araújo CP, Rosa CA. The influence of ripening period length and season on the microbiological parameters of a traditional Brazilian cheese. Braz J Microbiol 2013;44(3):743-749. [ Links ]
43. Çolaklar M, Taban BM, Aytaç SA, Barbaros H, Gürsoy A, Akçelik N. Application of bacteriocin-like inhibitory substances (BLIS)- Producing probiotic strain of Lactobacillus plantarum in control of Staphylococcus aureus in White-Brined cheese production. J AgrSci 2019;25(2019):401-408. [ Links ]
44. Bellio A, Astegiano S, Traversa A, Bianchi DM, Gallina S, Vitale N, et al. Behaviour of Listeria monocytogenes and Staphylococcus aureus in sliced, vacuum-packaged raw milk cheese stored at two different temperatures and time periods. Int Dairy J 2016; 57:15-19. [ Links ]
45. Stecchini MA, Sarais I, de Bertoldi M. The influence of Lactobacillus plantarum culture inoculation on the fate Staphylococcus aureus and Salmonella typhimurium in Montasio cheese. Int J Food Microbiol 1991;14(2):99-109. [ Links ]
46. Haines WC, Harmon LG. Effect of selected lactic acid bacteria on growth of Staphylococcus aureus and production of enterotoxin. Appl Microbiol 1973;25(3):436-441. [ Links ]
Received: December 14, 2021; Accepted: September 02, 2022











 text in
text in 



