Introduction
Streptococcus suis (S. suis) is an important pathogen in various countries in Europe, Asia, and the Americas. This bacterial species causes septicemia, meningitis, endocarditis, encephalitis, and bronchopneumonia in pigs and is a zoonotic agent1. Most pigs carry strains of multiple serotypes in their upper respiratory tract2,3. Symptoms of infection are often found in piglets from 2 wk of age but are more common in newly weaned pigs4. S. suis grows on blood agar plates (BAPs) and appears as small α-hemolytic colonies with a diameter of 0.5-1 mm. The colonies are grayish or transparent and are slightly mucoid. When using Gram staining, the cocci appear isolated, in pairs, and in short chains. S. suis is a facultatively anaerobic, immobile, and catalase- and oxidase-negative bacterium, which exhibits fermentative metabolism and produces acid from sugars, including mannitol. S. suis is also amylase positive, Voges-Proskauer negative, NaCl positive, and resistant to optochin; this bacterium has a capsule with epitopes that allow identification of 35 serotypes.
The identification of S. suis is performed by bacteriological methods, and it is recommended that biochemical tests be complemented with confirmatory serotyping. Serotyping is an important part of routine diagnostics and is based on capsular polysaccharide antigens found on the surface of S. suis strains, although some strains from clinical cases have been considered non-typable3. In the last 12 yr, more than 4,500 strains have been serotyped from diseased pigs worldwide; the serotypes that were isolated most frequently were, in decreasing order, 2 (28 %), 9 (19.4 %), and 3 (15.9 %), followed by serotypes 1/2 and 75. The distribution of serotypes isolated from infected pigs varies by geographic location6, but serotype 2 is isolated most frequently in meningitis cases from both pigs and humans7 .
The presumptive diagnosis of S. suis infection in piglets is often based on clinical signs and macroscopic lesions; confirmation is achieved by isolating the infectious agent and observing microscopic lesions in the affected tissues3. For clinical cases in pigs, isolation and identification of strains are relatively easy; successful identification can be achieved using a minimum of biochemical tests, and confirmation can be achieved by serotyping based on capsular polysaccharide antigens. However, the use of rapid, multitest biochemical kits may be misleading since some strains of S. suis can be misidentified as Streptococcus pneumoniae, S. bovis, and viridans group streptococci (e.g., S. anginosus and S. vestibularis). Likewise, although serotyping should be a part of routine identification of S. suis strains recovered from diseased pigs and humans to further confirm the pathogen’s identity, there may be cross-reactions between serotypes. Serotyping techniques are relatively simple; however, the production of antisera is laborious, time consuming, and expensive. In addition, there are strains that cannot be serotyped using antisera1. One disadvantage of serotyping with antisera is that non-encapsulated strains cannot be characterized and are called non-typable. These strains are identified using molecular techniques such as PCR as long as the genes of the capsule gene cluster (cps) have not been modified8.
Cloned the gene encoding glutamate dehydrogenase (gdh) from S. suis type 2, they observed that, similar to genes encoding other GDHs, the S. suis gene was highly conserved and had a very low mutation rate relative to that in other genes. With the help of gdh PCR, S. suis isolates can be identified. gdh PCR has been used as a quick and reliable diagnostic technique for samples from healthy and diseased animals, and it is also valuable in human cases7.
In the present study, characterization of S. suis strains isolated from sick animals, including serotype determination, was performed using molecular techniques, PCR of the gdh gene and multiplex PCR8,9. The goal of this study was to determine the serotypes of S. suis present in pig farms located in different areas of the Mexican Republic.
Material and methods
Microbiological sample collection and identification
Sixty-three (63) samples of organs were obtained from pigs with characteristic clinical signs of S. suis infection from different farms in the Mexican Republic. The samples were macerated in sterile phosphate-buffered saline, streaked onto 5% BAPs, and then incubated at 37 °C for 24 h. Colonies that were formed by α-hemolytic, Gram-positive cocci, grouped in chains, catalase and oxidase negative, were selected.
PCR of the gdh gene
DNA extraction was performed using the Wizard Plus SV minipreps DNA purification system (Promega, Madison, WI, USA), followed by gdh PCR to identify S. suis strains using an Invitrogen kit (Thermo Fisher Scientific, Carlsbad, CA, USA). The reactions were run on a Techne Progene thermal cycler under the following conditions: 2 min at 40 °C, 5 min at 94 °C, 35 cycles (1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C), and 7 min at 72 °C. The GenBank accession number of the sequence used to design the primers was AF229683.1. The sequence of the primers is JP4 (5´ GCAGCGTATTCTGTCAAACG 3´) y JP5 (5´CCATGGACAGATAAAGATGG 3´). The reaction products were visualized on a 2% agarose gel, stained with GelRed, using a Gene Genius bioimaging system (Artisan Technology Group, Champaign, IL, USA)9.
Two-step multiplex PCR
To determine serotypes, it was used a two-step multiplex PCR. First, a grouping PCR was carried out under the following thermal cycler conditions: 15 min at 95 °C, 30 cycles (30 sec at 94 °C, 90 sec at 90 °C, and 60 s at 72 °C), and 10 min at 72 °C. Second, a typing PCR was performed under the following thermal cycler conditions: 15 min at 95 °C, 30 cycles (30 sec at 94 °C, 90 sec at 53 °C, and 90 sec at 72 °C), and 10 min at 72 °C, using a Biometra thermocycler (Biometra, Göttingen, Germany). Qiagen multiplex PCR master mix was used, and the PCR products were visualized by electrophoresis on a 2% agarose gel (100 V, 40 min) stained with GelRed8. The primer sequences and predicted product size(s) for each type of PCR10 are showed in Table 1 .
Table 1 Primer sequences (5’ to 3’) used in this study and the predicted sizes of the PCR products [ Okura, et al 2014] 8
| Target cps group or type | Forward | Reverse | Size (bp) of products |
|---|---|---|---|
| I | TGGTTCAAATATCAATGCTC | ATTGGTTGTGAGTGCATTG | 933 |
| II | TCAAAATACGCACCTAAGGC | CACTCACCTGCCCCAAGAC | 823 |
| III | TGATTTGGGTGAGACCATG | CTCATGCTGGATAACACGT | 583 |
| IV | ACAGTCGGTCAAGATAATCG | TCAGCTTGGGTAATATCTGG | 455 |
| V | GGAAAGATGGAGGACCAGC | CCAACCAGACTCATATCCCC | 265 |
| III and VI | GATGCCCCAAGCGATATGCC | GGACCAACAATGGCCATCTC | 146 |
| GACGCACCAAGTGATATGCC | GGTCCGACAATAGCCATTTC | ||
| For PCR typing | Forward | Reverse |
Size (bp) of products |
| GROUP I | |||
| 3 | GGTTTTGATTGGTCTAGTTG | CTCTAAAGCTCGATATCTAC | 214 |
| 13 | TATGGTTAAAGGTGGAACTG | CCTTGTATATATTCCCTCCA | 408 |
| 18 | TAATGGGATAGTTGCGTTAC | ATACATAAAGTTGTCCTGCG | 617 |
| GROUP II | |||
| 2 and 1/2 | TTAGCAACGTTGCCAATAAG | AATCCTCCATTAAAACCCTG | 173 |
| 6 | GCTCACTATTTTTACATTACAC | TATTACTCCGCCAAATACAG | 278 |
| 1 and 14 | TTAGACAGACACCTTATAGG | CTAGCTTCGTTACTTGATTC | 386 |
| 16 | AAGGTTATCCACGAAAGATG | TCCGGCAATATTCTTTCAAG | 494 |
| 27 | AGACACTGCTTGCATTATTG | TCAGAATTACTTCCTGTTGC | 655 |
| GROUP III | |||
| 21 | TATCATATTGAGAATCTTCCC | TTGCGTAGCATACAAAGTTC | 160 |
| 28 | ATTATGTTGGTTGCAGAAGG | CGACTCAATTGTTGTAGTAG | 272 |
| 29 | TTCTGGGATTTTAGGAATGC | CATGAAATACGCACTTGTAC | 415 |
| 30 | TATTGCACTAGCTTCAGAAC | TGCATCCATAGTTGTATTCG | 568 |
| GROUP IV | |||
| 4 | GACTATCTGTATACCCAAAC | TCCTTCCAAGTATTCTCTAG | 903 |
| 5 | ATCTTAGGAATGATTCGGAC | ACCAGATATCTGAGCAAATG | 720 |
| 7 | AACTACCTACCTGAACTTTG | AGTCTAAAAGTGATCGAGTC | 566 |
| 17 | TAGCATCAGTTTATACGAGG | TAGTTTATCTGTGACACACC | 455 |
| 19 | GTGTCGCAAATCAAGTATTG | AAGCTAGTACAACAAGCATG | 348 |
| 23 | TAATGTATGCTCTGTCACTG | AACGAAACGGAATAGTTTGC | 221 |
| GROUP V | |||
| 8 | AAATAAGGTAGGAGCTACTC | ATCCAACCTTAGCTTTCTGT | 446 |
| 15 | ATCGTTTTGAGATTGAGTGG | TAAACGGATTCGGTTACTCA | 542 |
| 20 | TGTGGATTTCTGGGATAATC | TGTGGACGAATTACTACTTG | 698 |
| 22 | GCATTATCAGGATTCTTTCC | CCAATTGGGTGTTCAAAAAG | 296 |
| 25 | GTTTGCTCCGATCATAATAG | CCAGTAAAAGGACTCAATAC | 174 |
| GROUP VI | |||
| 9 | GAAAGTAGGTATATCTCAGC | GGGCTATTAAAACTCCTATC | 368 |
| 10 | TTTCCCATTTGCTTATGGAC | GGAATAAAAACGATTGGGAG | 633 |
| 11 | ATGCGATTGCAACAATTGAC | AGGCATGAGTAATACATAGG | 833 |
| 12 | AACAGGTATTTCAGGATTGC | CTCGGATAAAGATAATCAGC | 131 |
| 24 | TACTGAGATTTATTGGGACG | AAGCGATTGGATTACATTGC | 224 |
| 26 | TTATACCGAAATTTTGTTGCC | CGTCAATCATATAAAGTGGG | 472 |
| 33 | GATGTTTTCAACAGGTGTAC | CAAAGTACCTATTTTCAGCG | 710 |
| GROUP VII | |||
| 31 | ACAATCGTTTCTGCAATACG | GATGAAAACATCGTTGGTAG | 842 |
| 32 | AACCGCTGTTGAATTAAGAG | TTCGTTAGTTGAACTGTTCC | 570 |
| 34 | AAGTTTCATTCGAGGACTTC | GTATATAACACCGCAAGAAG | 246 |
| Internal control | |||
| 16S rRNA | GAGTTTGATCCTGGCTCAG | AGAAAGGAGGTGATCCAGCC | 1542-1553 |
Results
Microbiological sample collection and identification
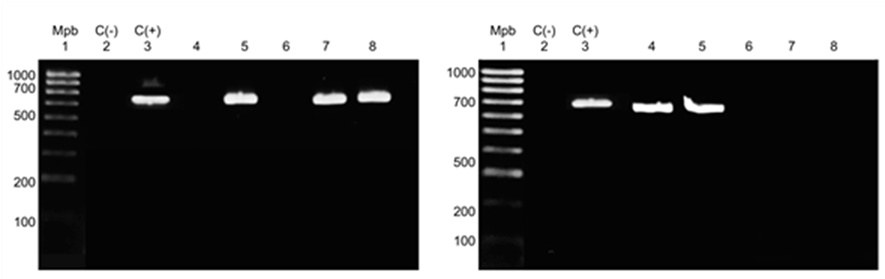
Of the 63 samples (lung, heart, and brain) collected from animals, 23 were positive for S. suis by PCR, with a product of 688 bp in length amplified by gdh PCR (Figure 1).
PCR of the gdh gene
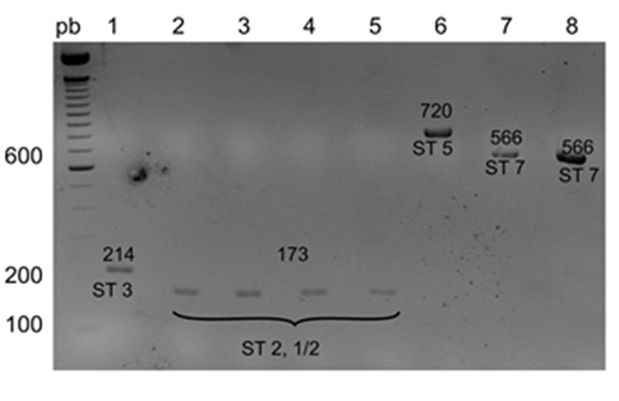
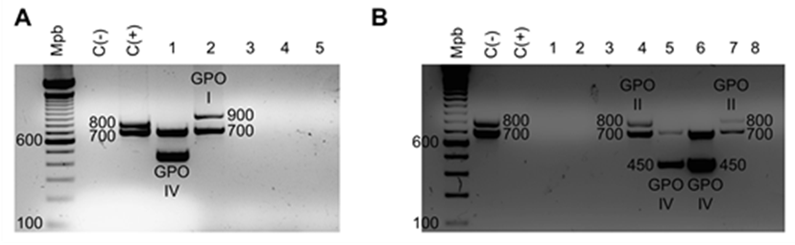
The results of the grouping PCR are shown in Figure 2; the samples were assigned to the following groups based on the sizes of the PCR products: I, II, IV, V, and VI, corresponding to the PCR product sizes of 933, 823, 455, 265, and 146 bp, respectively. Figure 3 shows the results of the typing PCR, wherein the serotype of each sample was determined. Serotypes 1/2 and 2 were characterized by the same size of their PCR products (173 bp); serotypes 3, 5, 7, 8, 9, 17, and 23 produced PCR fragments of 214, 720, 566, 446, 368, 445, and 221 bp, respectively. To differentiate serotypes 1/2 and 2, co-agglutination with specific antisera was used.
Gel A: lane Mbp, 100-1,000-bp marker; lane C(−), negative control; lane C(+), S. suis serotype 2; lanes 1 to 5, S. suis samples. Gel B: lane Mbp, 100-1,000-bp marker; lane C(−), negative control; lane C(+), S. suis serotype 2; lanes 1 to 8, samples.
Figure 2 Agarose gels (2%) showing results of the grouping PCR
Two-step multiplex PCR
The results of multiplex the PCR are shown in Table 2, which summarizes the information on the geographical areas where the samples were collected, the organs processed, and the serotypes identified.
Table 2 Results with the geographical area where the samples were collected, the processed organs and the serotypes
| Geographical area | Organ | Serotype |
|---|---|---|
| Perote-Veracruz | brain | 7 |
| Perote-Veracruz | brain | 7 |
| Perote-Veracruz | brain | 2 |
| Perote-Veracruz | brain | 2 |
| Perote-Veracruz | lung | 1/2 |
| Perote-Veracruz | brain | 3 |
| Perote, Veracruz | brain | 9 |
| Jalisco | lung | 9 |
| Jalisco | heart | 7 |
| Jalisco | lung | 9 |
| Jalisco | heart | 17 |
| Jalisco | heart | 9 |
| Jalisco | heart | 9 |
| Jalisco | lung | 3 |
| Jalisco | lung | 7 |
| Jalisco | heart | 23 |
| Jalisco | heart | 5 |
| Puebla | heart | 8 |
| Puebla | heart | 8 |
| Puebla | heart | 8 |
| Puebla | heart | 8 |
| Puebla | heart | 9 |
| La Piedad Michoacán | heart | 9 |
Discussion
PCR of the gdh gene is an attractive technique for use in both clinical laboratories and epidemiological studies9. Nevertheless, owing to the complexity of serotyping of S. suis strains8, have developed a two-step multiplex PCR. They sequenced and analyzed a group of genes from strains belonging to the 35 serotypes and reported that 31 of the serotypes (3 to 13 and 15 to 34) had specific genes, while serotypes 1 and 14 as well as 2 and 1/2 were almost identical. In the first step of the multiplex PCR, strains are classified into seven groups of cps genes, called homology groups (HGs). These cps genes are grouped within the same locus on the chromosome. Each HG is assigned a number (I-VII) and includes specific serotypes of S. suis. The typing PCR detects cps genes specific for each group and identifies the serotype. Molecular serotyping using multiplex PCR is attractive because animals are no longer required for the production of all 35 antisera since antisera are only needed to identify serotypes 1, 1/2, 2, and 1411. Previously, S. suis had been classified into 35 serotypes (1/2 and 1-34), and then, the number of serotypes was reduced to 33 because strains of serotypes 32 and 34 were re-classified as Streptococcus orisratti. More recently, it has been proposed to remove strains of serotypes 20, 22, 26, and 33 from the S. suis taxon12; however, in the present study, none of the isolated Mexican strains belonged to these serotypes.
In this study, there were determined serotypes of S. suis strains isolated from diseased animals from Mexican pig farms, and the diagnostic time was reduced, which was an advantage over routine diagnostic methods. It was found that serotype 9 was predominant in seven samples, isolated either from Jalisco (four samples), Veracruz, Michoacán, or Puebla (one sample each), followed by serotype 7, with four samples from Jalisco and Veracruz (two samples each), and serotype 8, with four samples from Puebla. Two samples from Veracruz were positive for serotype 2, and two samples from Jalisco and Veracruz were positive for serotype 3. One sample each was positive for serotypes 1/2, 5, 17, and 23. Jalisco was the state that presented the highest variation in serotypes, with six different lung and heart isolates belonging to serotypes 3, 5, 7, 9, 17, and 23. In Veracruz, five serotypes were detected in the lung and brain samples. Puebla was represented by two serotypes from heart samples. Only one serotype was detected in a heart sample from Michoacán.
Serotype assessment is a valuable tool for understanding the epidemiology of a particular outbreak, and serotyping also increases the success of vaccination programs within farms. It is possible that, this is the first report on the distribution of S. suis serotypes in areas dedicated to pig farming in Mexico. It was found that the predominant S. suis serotypes were comparable to those reported by Gottschalk et al10 between 2008 and 2011, who confirmed a relatively low prevalence of serotype 2 in North America compared with that in European and Asian countries. Among other serotypes, serotypes 1/2, 5, 9, and 14 have also been associated with outbreaks in pigs in North America and Europe10. Although it was not assess the prevalence, serotype 9 was the most frequent, followed by serotypes 7 and 8. These results support a previous hypothesis suggesting that a lower prevalence of S. suis serotype 2 is common for North American countries3. Since strains serotype 2 from North America and from Europe are genotypically and phenotypically different (with different virulence potential)13, it would be interesting to further study strains from Mexico to determine to which group of strains they belong. Similarly, it has been reported that serotype 9 strains from Europe are more homogenous and probably more virulent than North American strains14. Further evaluation of the Mexican strains is also needed to predict the level of pathogenicity of such strains. In addition, more studies, with a greater number of isolates from Mexico, are necessary to confirm this hypothesis. Although there is no clear association between serotypes and a given pathological condition, it has been reported that in Asian countries, strains isolated from diseased pigs primarily belonged to serotype 2, followed by serotypes 3, 4, 5, 7, 8, and 1/215. In some European countries, serotype 9 is most frequently recovered from diseased animals, followed by serotypes 1 and 14. However, in Canada, serotypes 1/2, 2, and 3 are the three most prevalent serotypes, followed by serotypes 4, 7, and 816. In humans, serotype 2 is the most prevalent serotype isolated, but serotypes 1, 4, 5, 14, 16, and 24 have also been reported5.
In South America, only two studies have been published, both from Brazil, which reported serotype 2 as the most prevalent, with a mean of 57.6 % of all cases, followed, in decreasing order of prevalence, by serotypes 1/2, 14, 7, and 9. Important pig-producing European countries, such as Denmark, Belgium, France, Germany, Italy, and the United Kingdom, have not recently reported the distribution of serotypes recovered from clinical cases in pigs. The latest reports from these countries published data on strains isolated between 1990 and 2000, and this lack of information is important. The only two countries with more recent data are Spain and the Netherlands. In fact, in Spain, serotype 2 is no longer the most prevalent serotype; it is now the second behind serotype 9, followed by serotypes 7, 8, and 3. In the Netherlands, serotype 9 was the most prevalent between 2002 and 2007, followed by serotypes 2, 7, 1 and 4. In studies conducted prior to 2002, serotype 1 appeared to be prevalent in countries such as Belgium and the United Kingdom1.
Conclusions and implications
The serotypes detected in the pig farming areas of Mexico included 1/2, 2, 3, 5, 7, 8, 9, 17, and 23. Jalisco was the state that presented the highest variation in serotypes, with six different serotypes. In Veracruz, five serotypes were detected in samples. Puebla was represented by two serotypes from samples. Only one serotype was detected in a sample from Michoacán. Serotype assessment is a valuable tool for understanding the epidemiology of a particular outbreak, and serotyping also increases the success of vaccination programs within farms. These findings are important for the characterization of serotypes present in Mexico and for outbreak prevention. To the best of our knowledge, this is the first report on the distribution of S. suis serotypes in areas dedicated to pig farming in Mexico. Research should continue to gain a better understanding of this microorganism and to have more complete data. The MALDI TOF MS is alternative methods for identify S. suis and the gdh test was considered specific for S. suis. The recN PCR is the test recognized as being specific for S. suis and so continue with the investigations.











 text in
text in