Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias pecuarias
On-line version ISSN 2448-6698Print version ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.11 n.4 Mérida Oct./Dec. 2020 Epub Mar 02, 2021
https://doi.org/10.22319/rmcp.v11i4.5111
Articles
Frequency of contamination and serovars of Salmonella enterica and Escherichia coli in an integrated cattle slaughtering and deboning operation
a Universidad Nacional Autónoma de México (UNAM). Facultad de Medicina Veterinaria y Zootecnia, Ciudad de México, México.
b UNAM. Facultad de Química. Ciudad de México. México.
c UNAM. Facultad de Medicina. Ciudad de México. México.
d Hospital General Dr. Manuel Gea González, Departamento de Ecología de Agentes Patógenos. Ciudad de México. México.
e Hospital Infantil de México Dr. Federico Gómez, Laboratorio de Bacteriología Intestinal. Ciudad de México. México.
This study aimed to determine the frequency of contamination and serovar diversity of Salmonella enterica (SE) and Escherichia coli (EC) in different stages of cattle slaughtering and deboning processes. Fecal, carcass, and primal cut (100 of each type) samples were collected in a Federally Inspected slaughterhouse in Mexicali, Baja California. EC was not analyzed in fecal samples because it is part of the gut microbiota. Strain identity was confirmed by biochemical methods and PCR, using the taxonomic genes invA and gadA for SE and EC, respectively. In EC, the presence of genes associated with the main pathotypes was also investigated. SE had a 34 % frequency in fecal samples, 3% in carcasses, and 2% in cuts, while Montevideo was the predominant serovar (72.5 % of the total strains). EC was detected in carcasses (34 %) and cuts (11 %) at an average concentration of 0.012 and 0.33 log CFU cm-2, respectively. Although several of the identified EC serovars were associated with enterotoxigenic or Shiga toxin-producing strains, none carried the virulence factors typically observed in these pathotypes. In summary, beef carcasses and cuts are not a relevant source of EC pathogenic strains. However, beef is an important reservoir of SE, which represents a public health risk. Genomic studies are required on the virulence profile and genes of SE strains commonly associated with subclinical infections and isolated from apparently healthy animals.
Key words Escherichia coli; Salmonella spp.; Cattle; Slaughter; Serovars; Pathotypes
El objetivo fue determinar la frecuencia de contaminación y la diversidad de serotipos de Salmonella enterica (SE) y Escherichia coli (EC) en diferentes etapas de los procesos de matanza y deshuese de bovinos. Se tomaron muestras de heces, canales y cortes primarios (100 de cada tipo) en un rastro Tipo Inspección Federal ubicado en Mexicali, Baja California. EC no se analizó en heces, por ser esta bacteria parte de la microbiota intestinal. La identidad de las cepas se confirmó por métodos bioquímicos y por PCR: genes taxonómicos invA y gadA, para SE y EC, respectivamente. En EC se investigó también la presencia de genes asociados con los principales patotipos. SE tuvo una frecuencia de 34 % en heces, 3 % en canales y 2 % en cortes, siendo Montevideo el serotipo predominante (72.5 % del total de cepas). EC se detectó en canales (34 %) y en cortes (11 %), en concentraciones promedio de 0.012 y 0.33 log UFC cm-2, respectivamente. Aunque varios de los serotipos de EC identificados están asociados con cepas enterotoxigénicas o productoras de toxinas tipo Shiga, ninguno portaba factores de virulencia característicos de estos patotipos. En resumen, las canales y cortes de bovino no son una fuente relevante de cepas patógenas de EC. En contraste, los bovinos constituyen un importante reservorio de SE, lo cual representa un riesgo para la salud pública. Se requieren estudios genómicos sobre el perfil de virulencia y los genes asociados con infecciones subclínicas en cepas de SE provenientes de animales aparentemente sanos.
Palabras clave Escherichia coli; Salmonella spp.; Bovinos; Sacrificio; Serotipos; Patotipos
Introduction
Intestinal infections caused by Salmonella enterica and the different pathotypes of E. coli, such as the Shiga toxin-producing E. coli (STEC), constitute a global public health problem1. Both pathogens are common contaminants of meat from different species, including beef2,3, which is the second most widely consumed meat type in Mexico4. Therefore, the characterization of circulating strains of S. enterica and E. coli in the beef cattle production chain is crucial to improve management of the risks associated with both pathogens.
In Mexico, most of the studies in this field focus on a single point in the production chain. For example, several authors have observed moderate frequencies (8 to 15 %) of Salmonella spp. in beef carcasses5-7, although the represented serovars are not reported in all cases. More comprehensive studies report higher levels of contamination (25 to 100 %) in hides, feces, lymph nodes, non-refrigerated carcasses, and meat samples8-10, as well as the predominance of certain serovars in some of the matrices analyzed. However, the comparison between studies is difficult due to variations in sample type, step of the production chain, method of analysis, geographical location, animal production system, and sanitary conditions of the studied process.
Regarding E. coli, the situation is similar. Most of the studies focus on a specific fragment of the production chain and deal with enterohemorrhagic STEC strains, such as E. coli O157:H76,11,12. Although previous studies have reported a low frequency (1 to 3%) of pathogenic E. coli strains in bovine carcasses and feces11-13, their distribution has not been thoroughly explored throughout the production chain. This information can contribute to identifying dissemination patterns in different processes and geographic regions, as well as measures to guarantee food safety and protect public health. Therefore, this study aimed to determine the frequency of contamination and serovar diversity of S. enterica and E. coli in a Federally Inspected slaughterhouse with horizontal integration of cattle slaughtering and deboning processes.
Material and methods
Study design and sample size determination
Samples were collected in three stages of the beef transformation process, from slaughtering to deboning: 1) Rectal contents collected after evisceration, 2) Hot carcasses, and 3) Primal cuts. Each stage was considered as an independent sampling, since it was not possible to determine in advance the destination of the animals, which were sold either as whole carcasses or as primal cuts. The sample size for each evaluated stage was calculated with the statistical equation used to determine the sample size of a population proportion when the number of elements in that population is unknown14:
With this formula it was obtained a sample size per stage of 96, which was rounded to 100, for a total of 300 samples in the study. The study was performed in September 2013 in an integrated beef production company in Mexicali, Baja California, comprising feedlots, slaughtering, and deboning operations. The sampled carcasses belonged to crossbred Bos indicus young bulls, with an average age of 24 to 30 months, originating from eight Mexican states and finished during an average of 190 days in the feedlot. The company was selected due to its level of integration, which allows having a production chain model in a single place. The slaughterhouse is 1 km off the feedlots and can process 300 heads of cattle per 8-h shift.
Sampling
Rectal contents
Fecal samples were collected from the rectum after evisceration, at 20 min postmortem. Approximately 100 g of fecal material were collected from each rectum. For this, viscera packages were momentarily held in the evisceration ramp. The rectum ligature was cut open and, using new nitrile gloves, it was collected the fecal samples in sterile bags, which were kept inside insulated containers with refrigerated gels (
Hot carcass sampling
Carcass sampling was conducted according to the methodology employed by the United States Department of Agriculture for the microbiological baseline studies for cattle15 with slight modifications. Instead of refrigerated carcasses, were sampled hot carcasses, and was used the peptone water from the same hot carcass to detect E. coli and S. enterica. Carcass swabs were collected from three different areas (leg, skirt, and brisket) of right halves. For that purpose, were used sponges pre-moistened with 10 ml of buffered peptone water and 10 x 10 cm2 sterile disposable frames (Meat/Turkey Carcass Sampling Kit, NASCO®, USA). The total sampling area per carcass was 300 cm2.
Microbiological analysis
Once the samples were taken, sponges were sealed in sterile plastic bags and kept inside insulated containers with refrigerant gels (
Biochemical identification
Salmonella strains were identified with substrates prepared in the laboratory according to the results of the following tests16: triple sugar iron (TSI); hydrogen sulfide, indole, and motility (SIM); Simmons citrate; urea; methyl red and Voges-Proskauer; malonate-phenylalanine; gluconate; arginine, ornithine, lysine, and control decarboxylase enzymes. Salmonella enterica subsp. enterica ser. Typhimurium ST19 was used as a positive control. This strain was obtained from the culture collection of the Hospital General Dr. Manuel Gea González, in Mexico City, isolated and characterized by VITEK 2 (bioMerieiux, France)17. The same tests were used for E. coli, except for amino acid decarboxylation16, using a strain of E. coli K12 was used as a positive control.
Molecular identification
Molecular identification was carried out by end-point PCR, using specific gene sequence primers typical of each species (Table 1). Genomic DNA was extracted from the purified strains previously refreshed in tryptic soy broth (MCD Lab®, PRONADISA-CONDA®, Spain) for 18 to 24 h using the “DNeasy Blood & Tissue Kit” (Qiagen, Inc., USA), following the instructions provided by the manufacturer. For S. enterica, was used the invA gene18, and for E. coli, the gadA gene19, which codes for the alpha subunit of the glutamate decarboxylase. Additionally, to identify the different pathotypes, was included six genes associated with enteropathogenic (EPEC), enterotoxigenic (ETEC), and Shiga toxin-producing (STEC) strains. Among these, the eaeA gene codes for an intimin, an important protein for adhesion through the translocated intimin receptor20. This gene is present in the genome of EPEC and STEC pathotypes. Moreover, the genes coding for Shiga toxins 1 (stx1) and 2 (stx2) usually occur in STEC strains, which show the same phenotype when they carry one or both of these genes20. It was also studied the presence of genes coding for the heat-stable (estA) and heat-labile (eltA) toxins associated with ETEC21 strains; as well as the bfp gene (bundle forming pilus), involved in adhesion to the intestinal epithelium, which is found in the genome of EPEC strains2. The PCR reactions were carried out in a total volume of 25 μl and the reagents from the Top Taq Master Mix Kit (QIAGEN®, USA) were used with the following final concentrations: 1.25 Units of Taq Polymerase, 1.5 mM of MgCl2, 1x PCR Buffer, 200 μM of each dNTP. The conditions used for each reaction were as described in previous publications (Table 1).
Table 1 Genes and primers used in the molecular characterization of Salmonella spp. and Escherichia coli
| Pathogen | Gene | Amplified fragment (bp) |
5´➜3´ Primer sequence | Ref. |
|---|---|---|---|---|
|
Salmonella spp. |
invA | 284 | 139 GTGAAATTATCGCCACGTTCGGGCAA 141 TCATCGCACCGTCAAAGGAACC |
(19) |
| gadA | 670 | gadA1: ACCTGCGTTGCGTAAATA gadA2: GGGCGGGAGAAGTTGATG |
(20) | |
| eaeA | 890 | EAE1: GTGGCGAATACTGGCGAGACT EAE2: CCCCATTCTTTTTCACCGTCG |
(21) | |
| stx1 | 582 | STX1F: ACACTGGATGATCTCAGTGG STX1R: CTGAATCCCCCTCCATTATG |
(21) | |
| E. coli | stx2 | 255 | STX2F: GGCACTGTCTGAAACTGCTCC STX2R: TCGCCAGTTATCTGACATTCTG |
(21) |
| estA | 190 | STa-F CTAATGTTGGCAATTTTTATTTCTGTA STa-R AGGATTACAACAAAGTTCACAGCAGTAA |
(22) | |
| eltA | 132 | LT-1 AGCAGGTTTCCCACCGGATCACCA LT-2 GTGCTCAGATTCTGGGTCTC |
(22) | |
| bfp | 324 | EP1, CAATGGTGCTTGCGCTTGCT EP2, GCCGCTTTATCCAACCTGGT |
(2) |
Amplified PCR products with high (gadA, eaeA, stx1) and low molecular weights (eltA and estA) were subjected to a 1% and 2% agarose gel electrophoresis (SeaKem® LE Agarose, Lonza, ME, USA), respectively. Gels were run in a tris/borate/EDTA buffer (TBE 1x) at 80 V for 50 min using SYBR Safe DNA Gel Stain (Invitrogen, USA) to reveal the DNA fragments. The visualization and digitization of images were performed in a Gel Logic 2200 imaging system (Kodak, USA) with the Care Stream® software (Carestream Health, Inc., USA). The same strains of both pathogens referred to in the biochemical identification were used as positive controls. Furthermore, to identify the E. coli pathotypes, we included strains of EPEC, ETEC, and STEC as controls. These strains were also obtained from the culture collection of the Hospital General Dr. Manuel Gea González and were previously characterized by VITEK 2.
Serotypification
Salmonella spp.
Serotypification of the somatic antigen (O). The serological identification of Salmonella strains was performed using the Kauffmann-White scheme22,23. The somatic antigen (O) was obtained by boiling the bacterial cultures (
Serotypification of the flagellar antigen (H). This antigen was obtained by inoculating the strains in a semisolid medium in Cragie's tubes and subculturing them in nutrient broth. Phase I and II H antigen were determined using the H antiserum Spicer-Edwards system (DIFCO) and monovalent sera (specific) from serogroups A, B, C, D, E, and F.
Although serovar determination was not carried out in a reference laboratory, the complete genome of the obtained strains was sequenced as part of another investigation24. This allowed to confirm, through in silico raw sequence analysis, the preliminary serotyping results and to determine the serovar of strains that were untypeable by biochemical methods.
E. coli
E. coli strains were serotyped by microagglutination in a 96-well microplate using 187 somatic antigen (O) antisera, and 53 flagellar antigen (H) antisera from rabbit (SERUNAM), following the method described by Ørskov and Ørskov25, with minor modifications.
Phylogroup classification. As certain E. coli phylogroups are associated with animals or humans, as well as with different bacterial pathotypes, it was decided to perform the classification into phylogenetic groups by PCR, according to the Clermont scheme26. This technique allows to divide the E. coli isolates into seven species-characteristic phylogenetic groups (A, B1, B2, C, D, E, and F) and one additional group, which corresponds to Cryptic Clade I. The test was performed by a quadruple PCR to detect the arpA, chuA, yjaA, and TSPE4.C2 genes. Moreover, when results suggest phylogroups E and C, an additional duplex PCR is performed for an allelic variant of the arpA (specific for group E) or trpA gene (specific for group C), including an internal control directed to the trpBA gene. Reactions were performed directly form fresh colonies grown for 24 h in TSA agar. The PCR reactions were performed in a 25 μl volume, under the same conditions previously described26. The PCR amplification products were subjected to a 2% agarose gel electrophoresis (SeaKem® LE Agarose, Lonza, USA) at 80 V for 50 min. Visualization and digitization of images were performed as previously described for S. enterica. E. coli K12 and representative strains of each phylogroup were included as positive controls. These strains were obtained from the culture collection of the Hospital General Dr. Manuel Gea González, previously classified according to the Clermont scheme26.
Results
From the analyzed samples (300 for Salmonella spp. and 200 for E. coli), it was obtained 84 isolates. Of these, 39 were identified as Salmonella spp. and 45 as E. coli, with a global frequency of 13.0 and 22.5 %, respectively. Only one carcass sample was positive for both bacteria.
Salmonella spp.
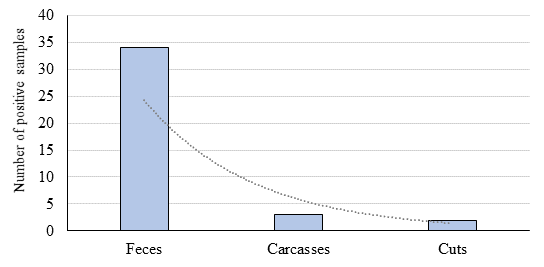
The frequency of contamination with Salmonella spp. was 34 % in fecal samples, and 3 and 2 % in carcasses and cuts, respectively (Figure 1). All these isolates were identified as Salmonella spp. by biochemical assays and PCR. Initially, it was identified two additional strains with positive results based on the biochemical tests and PCR, but they were untypeable by biochemical methods. However, when confirming the serovar by in silico raw sequence analysis, these two strains were identified as Pseudomonas putida, a species also carrying the invA gene27, and were therefore discarded.
The Chi-square test evidenced a strong association (
Table 2 Association between the frequency of contamination with x Salmonella spp. and Escherichia coli and sample type
| Sample type | n | Positivity % | Odds ratio | 95% C.I.1 |
|
P2 |
|---|---|---|---|---|---|---|
| Salmonella spp. | ||||||
| Feces | 100 | 34 | 20.1 | 7.5-53.5 | 58.5 | <0.0001 |
| Carcasses/cuts | 200 | 5 | ||||
| E. coli | ||||||
| Carcasses | 100 | 34 | 4.2 | 2.0-8.8 | 15.2 | <0.0001 |
| Cuts | 100 | 11 |
195% confidence interval for the odds ratio.
2Significance level (probability)
Regarding the serovars (Figure 2), it was possible to typifay 35 of the 39 isolates by serological methods. The remaining four strains were only partially characterized. Since they had a rough O antigen, it was only possible to obtain a partial antigenic formula based on the flagellar antigen. However, the in silico analysis, with raw reads from the fully sequenced genomes reported in another study24, allowed determining the serovar of 100% of the isolates. In total, it was identified five serovars: Bergen (n= 1), Reading (n= 2), Muenster (n= 3), Newport (n= 4), and Montevideo (n= 29). All Montevideo isolates were monophasic for the H antigen, although the antigenic formula allowed the identification of two subgroups within this serovar, 22 of them coming from feces, carcasses, and cuts, with the formula 6,7:g,m,s: -, while the seven remaining strains, all from fecal samples, had the formula 6,7:g,m,p,s:-.
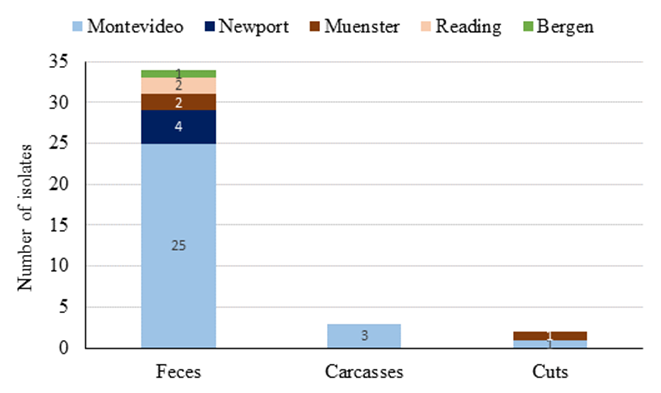
Figure 2 Distribution of Salmonella enterica subsp. enterica serovars according to isolation source (n=100 per sample type)
The serovar distribution per sample type showed Salmonella enterica subsp. enterica ser. Montevideo was present in all the analyzed matrices. Conversely, strains of Salmonella enterica subsp. enterica ser. Newport and Reading, detected at a lower frequency than Salmonella Montevideo, were only detected in fecal samples.
E. coli
E. coli was detected in 34 % of the carcasses and 11% of cuts. A strong association (
In cuts, the positive samples were distributed in a relatively uniform way, with five strains from isolated from the brisket, three from the skirt, and three from the leg. The concentration of this bacterium was low, both in carcasses and cuts, with values between 1 and 8 CFU cm-2. Of the 45 isolated and identified strains using 3M Petrifilm plates and CHROMAgar ECC, 41 showed a phenotype characteristic of the species. The four remaining strains, isolated from carcasses, showed atypical results; three were indole negative and slow lactose-fermenting, and one was positive for citrate, malonate, and cellobiose. However, all strains were molecularly confirmed by PCR, using the gadA gene as a taxonomic marker.
A total of 31 E. coli serovars were totally or partially identified (Table 3). The most frequent serogroups were O8 (29%) and O71 (19.4%), and the most common serovar was O1:H6 (9.7%).
Table 3 Frequency of Escherichia coli partially or totally identified serovars by sample type
| Sample type | n | Serovar |
|---|---|---|
| Carcass | 1 3 1 3 1 1 1 1 1 1 5 1 1 1 |
O28ab:- -:H30 -:H32 O1:H6 O113:- O154:H21 O156:- O166:H21 O32:- O6:- O8:- O8:H19 O8:H2 O8:H21 |
| Leg | 1 | -:H32 |
| Brisket | 1 1 2 1 |
O124:- O71:- O71:H12 O8:H8 |
| Skirt | 1 2 |
O7:H39 O71:H12 |
The predominant phylogenetic groups were A (60 %) and B1 (26.7 %), group B2 was absent, and groups C and D occurred at low frequencies (2.2 and 6.7 %). There were two strains with inconclusive results; therefore, they were not assigned to a phylogroup. It was interesting to observe how some serogroups were strongly associated with certain phylogenetic groups. In the serogroup O8, 8 out of 9 strains belonged to phylogroup B1. Similarly, 4 out of 5 strains in the serogroup O71 belonged to phylogroup A, and all strains from serogroup O1 belonged to phylogroup D (Figure 3).
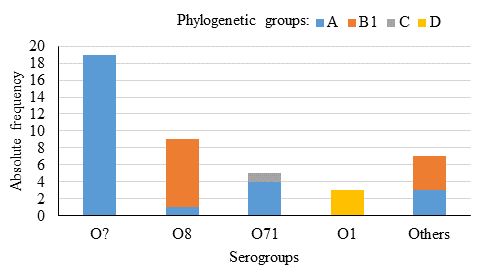
Figure 3 E. coli phylogenetic groups, based on the Clermont scheme26, represented in each of the identified serogroups (n=43)
Discussion
In several developed countries, with intensive beef production systems similar to those in developing countries, like Mexico, the contamination frequency of Salmonella spp. tends to be low in carcasses, meat, and feces28-30. However, in this study, it was observed a moderately high contamination frequency in feces, which coincides with previous reports in other Federally Inspected slaughterhouses in the country9. This indicates that, in Mexico, beef cattle farms may constitute an important reservoir of this pathogen. This surely represents an important challenge for the interventions applied during slaughter. Although the frequency of contamination in carcasses is drastically reduced compared to feces, a total control of the pathogen is not achieved. Furthermore, Salmonella was also detected in primal cuts, which shows the dissemination potential of this pathogen along the production chain. This is demonstrated by the detection of strains of the same serovar in feces, carcasses, and cuts. Additionally, these results are similar to those of previous studies (2 to 30 % positivity to Salmonella) in meat samples in supermarkets31,32, which only sell meat from Federally Inspected slaughterhouses. This situation implies a more complicated epidemiological situation in the commercialization chains associated with municipal slaughterhouses, which lack the infrastructure and sanitary conditions of those under federal inspection33. In fact, the positivity frequency to Salmonella in retail beef samples originated from municipal slaughterhouses generally exceeds 50 %10,34.
The above analysis shows the need to reinforce the control measures for Salmonella spp. in live animals, since the interventions applied on farms are limited. Therefore, the evaluation of the prevalence of Salmonella spp. in calves entering feedlots, the monitoring of infected animals and their separate management, and thus the detection of possible reservoirs are just some of the measures that could help to reduce the percentage of Salmonella carrier animals in slaughterhouses.
All the isolated serovars have been previously associated with human infections in Mexico35; therefore, the risk pose by these strains to public health should not be minimized. The clear predominance of Salmonella Montevideo in the evaluated processes is notorious and surprising, considering that the participating company fattens animals from eight Mexican states. The absence of previously common serovars in samples from Mexican beef cattle, such as Typhimurium, Anatum, and Agona, is also interesting36. Although previous studies have reported a variable distribution of Salmonella spp. serovars through time and between geographical areas and studies, the predominance of Salmonella Montevideo in this study is consistent with the increasing prevalence of this serovar in North America37,38. Furthermore, recent studies conducted in Mexico reported Montevideo and Reading serovars, but not Typhimurium, in strains isolated from cattle feces, carcasses, and lymph nodes8,9. In any case, it is difficult to determine the factors associated with the prevalence of specific strains in animal production systems without resorting to molecular studies to evaluate the genetic diversity of populations and the presence of genes associated with virulence, environmental persistence, and subclinical infections. However, these results indicate that apparently healthy cattle can carry Salmonella spp. at moderately high frequencies and that this pathogen can spread beyond the slaughtering process, with consequent risks to food safety.
The frequency of contamination with E. coli was similar to that of Salmonella in carcasses, and higher in cut samples; however, this bacterium occurred in low concentrations (<8 UFC cm-2). Although E. coli is part of the normal intestinal microbiota, the interventions applied in the slaughterhouse reduced three times the frequency of this bacterium in cut samples, in which the probability of finding positive samples was lower than in carcasses. Furthermore, in Mexico, the circulation of pathogenic strains in cattle appears to be lower than in other countries, such as the United States of America, where they are considered a public health problem39. This was further confirmed by the absence of the virulence factors associated with the STEC, EPEC, and ETEC pathotypes in the studied samples. Moreover, these findings coincide with previous studies13 that reported serovars (O157 and not-O157) associated with STEC strains (n=146), but only two of these carried the characteristic virulence factors. In Mexico, subsequent studies showed the same trend, reporting low rates (<1%) of contamination with pathogenic strains of E. coli in carcasses and ground beef9,40,41. This behavior could derive from multiple factors. Among these, the photoperiod, longer during the summer in the northernmost countries, has been considered responsible for the marked seasonal effect on the prevalence of pathogenic E. coli in cattle. Other authors have suggested that the circulation of different enterobacteria, with cross-reaction of somatic (O) antigens, could be a negative selection factor of E. coli pathotypes in Mexican cattle populations42. This is in line with the high percentage of serum samples, from apparently healthy cattle, with a bactericide response against E. coli O157 (71 %), in herds from central Mexico43.
Regarding the identified E. coli phylogroups, the predominance of A and B1 is similar to what is commonly observed in strains of animal origin44,45. In line with the absence of virulence genes associated with pathotypes, only one strain was classified in group C, to which other STEC strains of animal origin belong44,46. However, practically all the identified serovars have been associated with the STEC or ETEC pathotypes, which are important in foodborne diseases47-49. The potential health risks of non-pathogenic strains should not be overlooked since they could acquire virulence factors by incorporating plasmids or phages50,51. Hence, further research is needed in this area.
Conclusions and implications
This study shows that nearly one third of the cattle approved for slaughter carry different serovars of Salmonella enterica in their feces, despite being apparently healthy animals. Furthermore, the results show the ability of the pathogen to spread to the following segments of the production chain, with the consequent risks to public health. Hence, it is important to conduct further studies on the genetic factors of S. enterica associated with the establishment of subclinical infections in cattle and their persistence in livestock populations. Moreover, the results for E. coli show, as in other regions of the country, a low circulation of pathogenic strains of E. coli in beef carcasses and cuts. However, the analyzed samples were obtained from a single slaughterhouse, and the scope of this study, for E. coli, does not consider hide or feces samples, in which the probability of finding pathogenic strains is higher.
Acknowledgments and conflicts of interest
This study was carried out with resources from the SAGARPA-CONACYT sector fund, project 109127. We appreciate the technical support of the professionals in the Hospital General “Dr. Manuel Gea González”, the School of Medicine of the Universidad Nacional Autónoma de México, and the Universidad Autónoma de Baja California, for their assistance in conducting the experiments and the laboratory analyses. The authors declare that they have no conflicts of interest regarding this publication.
REFERENCES
1. WHO. WHO estimates of the global burden of foodborne diseases. Foodborne disease burden epidemiology reference group 2007-2015. World Health Organization. http://www.who.int/foodsafety/areas_work/foodborne-diseases/ferg/en/. Accessed Oct 25, 2018. [ Links ]
2. Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013;26(4):822-880. [ Links ]
3. Koohmaraie M, Scanga JA, De La Zerda MJ, Koohmaraie B, Tapay L, Beskhlebnaya V, et al. Tracking the sources of Salmonella in ground beef produced from nonfed cattle. J Food Protect 2012;75(8):1464-1468. [ Links ]
4. FAOSTAT. Food Balance Sheets. Food and Agricultural Organization of the United Nations, Statistics Division. http://faostat3.fao.org/download/FB/FBS/E . Consultado 25 Feb, 2019. [ Links ]
5. Varela-Guerrero JA, Talavera-Rojas M, Gutiérrez-Castillo AC, Reyes-Rodríguez NE, Vázquez-Guadarrama J. Phenotypic-genotypic resistance in Salmonella spp. isolated from cattle carcasses from the north central zone of the State of Mexico. Trop Anim Health Prod 2013;45(4):995-1000. [ Links ]
6. Hernández-San Juan S, Zúñiga-Estrada A, Sánchez-Ortega I, Castro-Rosas J, Román-Gutiérrez AD, Santos-López EM. Condiciones microbiológicas en el proceso de sacrificio en un rastro municipal del estado de Hidalgo, México. Vet Mex 2007;38(2):187-194. [ Links ]
7. Pérez-Montaño JA, González-Aguilar D, Barba J, Pacheco-Gallardo C, Campos-Bravo CA, García S, Heredia NL, Cabrera-Díaz E. Frequency and antimicrobial resistance of Salmonella serovars on beef carcasses at small abattoirs in Jalisco State, Mexico. J Food Prot 2012;75(5):867-873. [ Links ]
8. Gragg SE, Loneragan GH, Nightingale KK, Brichta-Harhay DM, Ruiz H, Elder JR, et al. Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl Environ Microbiol 2013;79(15):4744-50. [ Links ]
9. Narvaez-Bravo C, Miller MF, Jackson T, Jackson S, Rodas-Gonzalez A, Pond K, Echeverry A, Brashears MM. Salmonella and Escherichia coli O157:H7 Prevalence in cattle and on carcasses in a vertically integrated feedlot and harvest plant in Mexico. J Food Prot 2013;76(5):786-795. [ Links ]
10. Zaidi MB, McDermott P, Fedorka-Cray P, Leon V, Canche C, Hubert S, et al. Nontyphoidal Salmonella from human clinical cases, asymptomatic children and raw retail meats in Yucatan, Mexico. Clin Infect Dis 2006;42:21-28. [ Links ]
11. Callaway TR, Anderson RC, Téllez G, Rosario C, Nava GM, Eslava C, et al. Prevalence of Escherichia coli O157 in cattle and swine in central Mexico. J Food Prot 2004;67(10):2274-2276. [ Links ]
12. Amézquita-López BA, Quinones B, Cooley MB, Leon-Felix J, Castro-del Campo N, Mandrell RE, et al. Genotypic analyses of shiga toxin-producing Escherichia coli O157 and non-O157 recovered from feces of domestic animals on rural farms in Mexico. PLoS One 2012;7(12):e51565. [ Links ]
13. Varela-Hernández JJ, Cabrera-Díaz E, Cardona-López MA, Ibarra-Velázquez LM, Rangel-Villalobos H, Castillo A, et al. Isolation and characterization of Shiga toxin-producing Escherichia coli O157:H7 and non-O157 from beef carcasses at a slaughter plant in Mexico. Int J Food Microbiol 2007;113(2):237-41. [ Links ]
14. Jekel JF, Katz DL, Elmore JG. Epidemiology, biostatistics, and preventive medicine. Third ed. Philadelphia, PA, United States of America: SAUNDERS ELSEVIER; 2007. [ Links ]
15. USDA/FSIS. Nationwide Sponge Microbiological Baseline Data Collection Program: Cattle. https://www.fsis.usda.gov/wps/wcm/connect/197ddb9c-1fa5-411e-ab3f-3d253c879370/Baseline_Data_Cattle.pdf?MOD=AJPERES . Accessed Mar 5, 2019. [ Links ]
16. Barrow GI, Feltham RKA. Cowan and Steel's manual for the identification of medical bacteria. 3rd Ed. ed. Cambridge, United Kingdom: Cambridge University Press; 1993. [ Links ]
17. Bruins MJ, Bloembergen P, Ruijs GJHM, Wolfhagen MJHM. Identification and susceptibility testing of Enterobacteriaceae and Pseudomona aeruginosa by direct inoculation from positive BACTEC blood culture bottles into Vitek 2. J Clin Microbiol 2004;42(1):7-11. [ Links ]
18. Rahn K, De Grandis SA, Clarke RC, Curtiss R, Gyles CL. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 1992;6:271-279. [ Links ]
19. McDaniels AE, Rice EW, Reyes AL, Johnson CH, Haugland RA, Stelma GNJ. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β-d-glucuronidase. Appl Environ Microbiol 1996;62(9):3350-3354. [ Links ]
20. DebRoy C, Roberts E. Screening petting zoo animals for the presence of potentially pathogenic Escherichia coli. J Vet Diagn Invest 2006;18(6):597-600. [ Links ]
21. Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog 2011;3(9). [ Links ]
22. Grimont PAD, Weill F. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur. https://www.pasteur.fr/sites/default/files/veng_0.pdf . Accessed Feb 25, 2019. [ Links ]
23. Popoff MY, Le Minor LE. Salmonella. In: Bergey's Manual of Systematics of Archaea and Bacteria. Whitman FR, et al editors. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2015:764-799 [ Links ]
24. Delgado-Suárez EJ, Selem-Mojica N, Ortiz-López R, Gebreyes WA, Allard MW, Barona-Gómez F, Rubio-Lozano MS. Whole genome sequencing reveals widespread distribution of typhoidal toxin genes and VirB/D4 plasmids in bovine-associated nontyphoidal Salmonella. Sci Rep 2018;8(1):9864. [ Links ]
25. Ørskov F, Ørskov I. Serotyping of Escherichia coli. Method Microbiol 1984;14:43-112. [ Links ]
26. Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylogroups. Environ Microbiol Rep 2013;5(1):58-65. [ Links ]
27. Duan J, Jiang W, Cheng Z, Heikkila JJ, Glick BR. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS One 2013;8(3):e58640. [ Links ]
28. Eguale T, Engidawork E, Gebreyes WA, Asrat D, Alemayehu H, Medhin G, Johnson RP, Gunn JS. Fecal prevalence, serovar distribution and antimicrobial resistance of Salmonellae in dairy cattle in central Ethiopia. BMC Microbiol 2016;16:20. [ Links ]
29. Sandt CH, Fedorka-Cray PJ, Tewari D, Ostroff S, Joyce K, M'Ikanatha NM. A comparison of non-typhoidal Salmonella from humans and food animals using pulsed-field gel electrophoresis and antimicrobial susceptibility patterns. PLoS One 2013;8(10):e77836. [ Links ]
30. Khen BK, Lynch OA, Carroll J, McDowell DA, Duffy G. Prevalence and characteristics of Salmonella in the beef chain in the Republic of Ireland. Zoonoses Public Health 2014;61(8):534-6. [ Links ]
31. Ballesteros-Nova N, Rubio-Lozano MS, Delgado-Suárez EJ, Méndez-Medina RD, Braña-Varela D, Rodas Suárez O. Perfil de resistencia a antibióticos de serotipos Salmonella spp. aislados de carne de res molida en la Ciudad de México. Salud Pública México 2016;58(3):1-7. [ Links ]
32. Rubio-Lozano MS, Martínez-Bruno JF, Hernández-Castro R, Bonilla-Contreras C, Méndez MRD, Núñez-Espinosa JF, et al. Detección de Listeria monocytogenes, Salmonella y Yersinia enterocolitica en carne de res en puntos de venta en México. Rev Mex Cienc Pecu 2013;4(1):107-115. [ Links ]
33. Signorini M. Evaluación de riesgos de los rastros y mataderos municipales. Nacameh 2007;1(2):118-141. [ Links ]
34. Cabrera-Díaz E, Barbosa-Cárdenas CM, Pérez-Montano JA, González-Aguilar D, Pacheco-Gallardo C, Barba J. Occurrence, serovar diversity, and antimicrobial resistance of Salmonella in ground beef at retail stores in Jalisco state, Mexico. J Food Prot 2013;76(12):2004-2010. [ Links ]
35. Gutiérrez-Cogco L, Montiel-Vázquez E, Aguilera-Pérez P, González-Andrade MC. Serotipos de Salmonella identificados en los servicios de salud de México. Salud Publica Mex 2000;42(6):490-495. [ Links ]
36. Zaidi MB, Calva JJ, Estrada-García MT, León V, Vázquez G, Figueroa G, et al. Integrated food chain surveillance system for Salmonella spp. in Mexico. Emerg Infect Dis 2008;14(3):429-435. [ Links ]
37. Habing GG, Manning S, Bolin C, Cui Y, Rudrik J, Dietrich S, Kaneene JB. Within-Farm Changes in dairy farm-associated Salmonella subtypes and comparison to human clinical isolates in michigan, 2000-2001 and 2009. Appl Environ Microbiol 2015;81(17):5724-35. [ Links ]
38. Webb HE, Brichta-Harhay DM, Brashears MM, Nightingale KK, Arthur TM, Bosilevac JM, et al. Salmonella in peripheral lymph nodes of healthy cattle at slaughter. Front Microbiol 2017;8:2214. [ Links ]
39. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, et al. Foodborne illness acquired in the united states-major pathogens. Emerg Infect Dis 2011;17(1):7-15. [ Links ]
40. Gómez-Aldapa CA, Díaz-Cruz CA, Cerna-Cortés JF, Torres-Vitela MR, Villarruel-López A, Rangel-Vargas E. Escherichia coli O157 in ground beef from local retail markets in Pachuca, Mexico. J Food Prot 2013;76(4):680-684. [ Links ]
41. Gallegos M, Morales A, Álvarez G, Vázquez J, Morales L, Martínez I, Maldonado J. Caracterización de aislados de Escherichia coli O157:H7 en canales de bovinos y porcinos mediante PCR. Revista Científica, FCV-LUZ 2009;XIX(2):139-146. [ Links ]
42. Chart H, Okubadejo A, Rowe B. The serological relationship between Escherichia coli O157 and Yersinia enteocolitica O9 using sera from patients with brucellosis. Epidemiol Infect 1992;108(1):77-85. [ Links ]
43. Navarro A, Eslava C, García de la Torre G, León LA, Licona D, León L, Zarco LA, Cravioto A. Common epitopes in LPS of different Enterobacteriaceae are associated with an immune response against Escherichia coli O157 in bovine serum samples. J Med Microbiol 2007;56:1447-1454. [ Links ]
44. Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol 2011;11(3):654-662. [ Links ]
45. Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MI, Gomes TA, Amaral LA, Ottoboni LM. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol 2010;1(10):161. [ Links ]
46. CDC. Center for Disease Control and Prevention. Multistate outbreak of shiga toxin-producing Escherichia coli O145 infections (final update). https://www.cdc.gov/ecoli/2012/o145-06-12/index.html . Accessed Mar 5, 2019. [ Links ]
47. Blanco M, Blanco JE, Mora A, Dhabi E, Alonso MP, González EA, et al. Serovars, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J Clin Microbiol 2004;42(2):645-651. [ Links ]
48. Bettelheim KA. The Non-O157 Shiga-Toxigenic (Verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit Rev Microbiol 2007;33(1):67-87. [ Links ]
49. De Moura C, Ludovico M, Valadares GF, Gatti MSV, Leite DS. Detection of virulence genes in Escherichia coli strains isolated from diarrheic and healthy feces of dairy calves in Brazil. Arq Inst Biol 2012;79(2):273-276. [ Links ]
50. Martínez-Castillo A, Muniesa M. Implications of free Shiga toxin-converting bacteriophages occurring outside bacteria for the evolution and the detection of Shiga toxin producing Escherichia coli. Front Cell Infect Microbiol 2014;4:46. [ Links ]
51. Hallewell J, Niu YD, Munns K, McAllister TA, Johnson RP, Ackermann HW, Thomas JE, Standford K. Differing populations of endemic bacteriophages in cattle shedding high and low numbers of Escherichia coli O157:H7 Bacteria in feces. Appl Environ Microbiol 2014;80(13):3819-3825. [ Links ]
Received: October 16, 2018; Accepted: September 23, 2019











 text in
text in