Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias pecuarias
On-line version ISSN 2448-6698Print version ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.10 n.1 Mérida Jan./Mar. 2019
https://doi.org/10.22319/rmcp.v10i1.4512
Articles
Isolation and identification of potentially probiotic lactic acid bacteria for Holstein calves in the Mexican Plateau
a Colegio de Postgraduados, Campus Montecillo. Km 36.5 Carretera México Texcoco. Montecillo Estado de México. CP 56230.
b Benemérita Universidad Autónoma de Puebla. Facultad de Medicina.
Neonate calves are continuously exposed to a wide range of microorganisms in the environment, including diarrhea-causing enteropathogens. Lactic acid bacteria (LAB) was isolated from the oral mucosa of calves, and colostrum and milk from Holstein cows, the strains identified and their resistance to acid pH and bile salts tested. Isolation was done on plated de Man-Rogosa-Sharpe agar. Once decontaminated, the LAB colonies were morphologically and biochemically characterized. Sixteen of the isolated bacterial strains were selected: 12 from oral mucosa, 2 from milk and 2 from colostrum. After testing for resistance to an acid environment (pH 4 and 4.5) and bile salts (0.3 and 1.5 g), the five most resistant species (pH 4 and 1.5 g bile salts) were identified with the API 50 CHL system: Leuconostoc mesenteroides, Pediococcus pentosaceus, Lactobacillus plantarum, Lactobacillus crispatus and Lactococcus lactis. These strains have probiotic potential in calves.
Key words: Calves; Probiotics; Isolation; Resistance
Los becerros neonatos son expuestos continuamente a un amplio rango de microorganismos habitantes del ambiente, y a patógenos causantes de diarrea. El objetivo de este estudio fue aislar e identificar bacterias acido lácticas (BAL) de la mucosa oral de becerros, calostro y leche de vacas Holstein. El aislamiento de BAL se realizó en caldo y placas con medio ManRogosa Sharp (MRS). Una vez purificadas las colonias bacterianas, se realizaron pruebas morfológicas y bioquímicas para la caracterización de BAL. Se aislaron 16 colonias bacterianas, de las cuales se clasificaron en 12 de la mucosa oral, 2 en leche y 2 en calostro. Estas colonias se evaluaron a pH ácido (4.0 y 4.5) y sales biliares (SB: 0.3 y 1.5 g). Posteriormente se identificaron con el Sistema API50CHL. Las especies aisladas con resistencia a pH de 4 y 1.5 de SB fueron: Leuconostoc mesenteroides, Pediococcus pentosaceus, Lactobacillus plantarum, Lactobacillus crispatus y Lactococcus lactis. Estas colonias cuentan con un potencial probiótico para ser usadas en becerros.
Palabras clave: Becerras; Probióticos; Aislamiento; Resistencia
Introduction
A number of problems can arise when breeding heifers for replacement, including poor colostrum supply, feeding with low quality milk substitutes and sudden changes in ration1. These substandard breeding practices can lead to diarrhea, caused mainly by enteropathogens, with mortality rates exceeding 10 % during the first weeks of life2. Antibiotics are used to reduce mortality, but many pathogenic strains have developed resistance, negatively affecting animal health3,4. Several veterinary pharmaceutical laboratories now promote the use of probiotics containing lactic acid bacteria (LAB) and claim benefits such as prevention and reduction of diarrhea, and improved weight gain. However, to qualify as efficient probiotics these products must comply with certain requirements. For example, the minimum number of microorganisms required in a calf’s intestine to generate adequate health is 106 colony-forming units (CFU)/ml1.
Clinical trials done over the last ten years have found that 45% of probiotics on the market contain LAB with null efficiency in the prevention of diarrhea in heifers. Some even seemed to aggravate diarrhea incidence and severity4,5, and provided no improvements in daily weight gain and feed conversion6,7. The same still holds true for the probiotics marketed to dairy cattle production units: low viability probiotic microorganisms are used, and bacteria species other than those on the label have been identified8. Some strains come from different geographical regions and/or other animal species, which causes low viability and probiotic activity4. The present study objective was to isolate and identify bacteria with probiotic potential (i.e. resistance to acid pH and bile salts) in Holstein cattle in the Plateau region of Mexico.
Material and methods
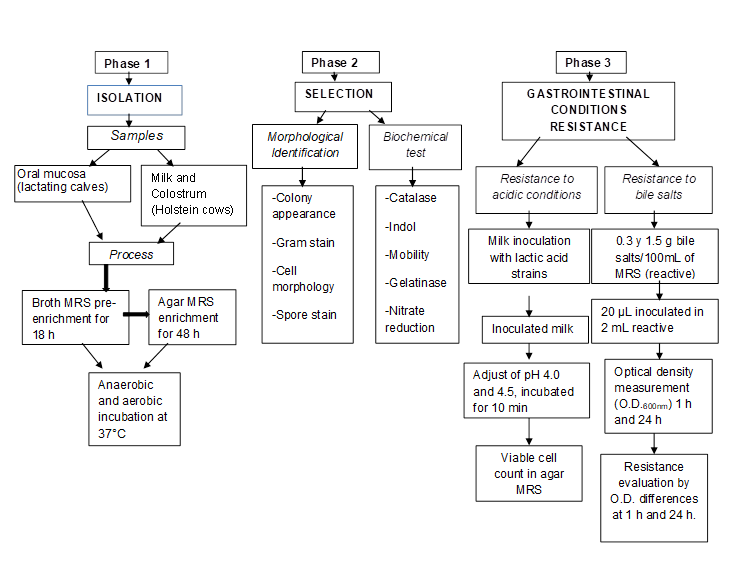
Isolation of bacteria from oral mucosa and colostrum
The experimental animals consisted of five lactating calves (30 d of age) and five multiparous adult cows in lactation, all Friesian Holstein from College of Postgraduates (Colegio de Postgraduados, Campus Montecillo) installations. Colostrum samples were taken from five newly-calved Holstein cows on the private Xalapango ranch. Both sites are located in the Texcoco Valley, in the State of Mexico, Mexico (18°21’ and 20°17’ N; 98°36’ and 100°36’ W)9.
Sampling
Oral mucosa: Duplicate exudate samples were taken of the oral mucosa from each lactating calf, by rubbing the mucosa for 3 sec with a swab (3MTMSwab-sampler) prior to the morning feeding. Each swab was placed inside a sterile tube with 10 ml buffered peptone water (RS96010BPW).
Colostrum and milk: Before sampling, the cows’ nipples were cleaned, disinfected and pulled down, and 5 ml of colostrum and 10 ml of milk collected per group of cows. Samples were deposited in sterile vials, kept at 4 °C and immediately transferred to the laboratory for analysis following standardized procedures10.
Sample processing
The samples were pre-enriched (to favor LAB growth) in liquid culture medium11. Duplicate samples (1 ml) of the oral mucosa bacterial suspension were taken and placed in tubes containing 5 ml de Man/Rogosa/Sharp (MRS) broth. The inoculated tubes were divided into two random groups: one under aerobic conditions and the other under anaerobic conditions, both were incubated at 37 °C for 18 h. In the colostrum and milk samples, 200 μl were taken in duplicate and deposited in tubes containing 5 ml MRS broth, and kept in a desiccator under an anaerobic environment (induced by a burning candle) for 18 h at 37 °C. Samples were then taken from the tubes with an inoculation loop and sown in Petri dishes containing MRS agar12, and incubated at 37 °C for 48 h under anaerobic and aerobic conditions. A strain of Lactobacillus casei ATCC was used as a positive control and one of E. coli O42 as a negative control (both donated by the Universidad Autónoma de Querétaro).
Bacteria selection
During the sowing process the cultures were seriated according to sample duplication in the solid MRS medium. The result was a total of 54 colonies with LAB characteristics based on colony size, shape, surface, elevation, edge and color13. Strain characterization was done by the Gram stain test, cell morphology, spore staining and the catalase test12. Indole production and motility tests were done in SIM (hydrogen sulfide, indole, motility) culture medium; gelatin hydrolysis and nitrate reduction tests were also done14. A second selection of the isolated colonies was made based on the best scores and ideal coccobacilli and bacilli morphology. A total of 27 colonies were identified which were cultured in 5 ml MRS broth for 18 h for later evaluation as probiotic bacteria. Duplicate 800 µl samples were taken from each bacterial suspension and transferred to Eppendorf tubes containing 800 µl sterile 50% glycerol as a cryoprotectant. These were stored at -20 ºC for 3 h and subsequently at -80 ºC indefinitely.
Resistance and survival of selected strains under gastrointestinal conditions
Resistance to acid pH
Inoculum preparation: A further selection was made of the 27 colonies obtained in the isolation process; 16 were chosen for having well-defined coccobacilli and bacilli morphology. Of these sixteen, twelve were isolates from oral mucosa, two from milk and two from colostrum. All selected colonies were reactivated by raising storage temperature to -20 ºC and then to room temperature. Each colony was then transferred into tubes containing 5 ml MRS broth and incubated at 37 ºC under anaerobic conditions for 24 h. A 1 ml sample of each bacterial suspension (106 Log10 CFU/ml) was added to tubes containing 9 ml MRS broth and incubated another 18 h. These final bacterial suspensions were centrifuged at 2,056 xg for 10 min, and the cellular packages resuspended in 10 ml sterilized (110 °C for 15 min) skim milk (Alpura® 2000®) for the resistance test at pH 4.5 and 4.0. The milk functioned as a protective medium and a vehicle for probiotic microorganisms15, following the protocol described by Fernández de Palencia et al16. Resistance to acid pH conditions was assessed by reducing the pH to which the bacterial cells were exposed. Reduction of pH was done with controlled HCl aliquots. When PH stabilized at 4.5 and 4.0, samples were incubated at 37 °C for 10 min. Subsequently, 1 ml of each suspension was taken to make serial dilutions. From each dilution, 100 µl was taken and sown in MRS agar to estimate bacterial cell viability. The colonies evaluated at pH 4.5 and 4.0 were sown at 10-6 and 10-7 in a milk suspension.
Resistance to bile salts exposure in microtitre plates
The selected colonies were exposed to bile salts (BS) in microtitre plates (BD PrimariaTM) with 3.5 ml wells. One plate was used per concentration. Before beginning the test, two flasks were prepared containing 100 ml MRS broth: one with 0.3 g bovine BS and the other with 1.5 g bovine BS (Oxgall DifcoTM)17,18. Two negative controls were run: MRS with BS and without bacteria, and MRS without BS and with bacteria. The BS resistance test was run in triplicate and each colony occupied six plate wells. In addition, 2 ml BS solution (MRS with BS and without bacteria) and 20 μl (1:10 v/v) of bacterial suspension were incubated for 18 h. One hour after inoculation and before the plates were incubated, the optical density (OD) of each suspension was measured at 600 nm using a spectrophotometer (GENESYS 10 UV/Thermo Spectronic). The OD reading was taken again once the plates had been incubated under anaerobic conditions at 37 ºC for 24 h.
Biochemical identification and collection of strains with probiotic potential
Colonies with LAB characteristics were identified using the APICHL system (BioMerieux SA, France). In this procedure the colonies were reactivated in 5 ml MRS broth under anaerobic conditions at 37 ºC for 18 H, adding 50CH diluent (supplied with the gallery: API50CH) following manufacturer instructions (see Figure 1 for summary of procedure). The prepared suspension was added to 50 microtubes in the gallery, and the domes of these microtubes filled with sterile mineral oil to generate anaerobic conditions. The inoculated galleries (one per colony) were kept at 37 ºC for 48 h to establish each colony’s biochemical profile. Results interpretation was done based on color change in the API50CHL medium of each microtube: blue is negative, and yellow and black indicate positive values (plate safety sheet). Results were analyzed with the Apiweb® computer system.
Statistical Analyses
When analyzed with Kolmogorov-Smirnov test the data exhibited a normal distribution, and a Levene test showed variance homogeneity. Means were compared with an ANOVA and a Tukey test; significance level was 0.05%. All analyses were run with the SPSS ver. 15 statistics package19.
Results and discussion
Colony isolation and growth
The colonies cultured in the anaerobic environment exhibited better growth than those cultured under aerobic conditions. Based on morphology, the selected colonies were Gram positive coccobacilli and bacilli, with no spores, catalase negative, no motility, no indole or gelatinase production and nitrate reduction negative.
The colonies from the oral mucosa samples had an average size of 2 to 4 mm in diameter with homogeneous morphological characteristics; circular shape, convex elevation, complete edge, smooth surface and white color without pigments. Of the milk samples only 20 % supported bacterial colony growth, which had an average size of 2.5 mm diameter. Those colonies isolated from the colostrum were beige in color and varied in size from 1 to 5 mm diameter. Probiotic bacteria have generally been isolated from the oral, vaginal, and intestinal mucosa of healthy calves and from milk samples11,20. Lactobacilli colonies isolated from the oral mucosa and milk have the capacity to adapt and survive13. This is due to the presence of the hemin group, which allows them to activate the respiratory chain with oxygen as the electron recipient21. The different LAB genera share morphological, metabolic and physiological characteristics such as shape, elevation, edge, color and biochemical reactions13. For the purpose of probiotic strain selection, their cell morphology and biochemical tests have been reported as basic5,22, although it is recommended that selection be complemented with molecular studies23.
API biochemical strain identification
Colony identification based on carbohydrate fermentation profile (API50CHL-BioMerieux) produced a 96 to 99 % effectiveness interval (Table 1). Identified colonies from the oral mucosa included six Lactobacillus, five Leuconostoc and one Pediococcus, while those from the colostrum included Leuconostoc and Lactobacillus.
Table 1: Colony counts in de Man-Rogosa-Sharpe (MRS) agar immediately after exposure to acid pH, and biochemical strain identifications
| Strain count |
T1 pH 6.5 |
T2 pH 4.5 |
T3 pH 4.0 |
Biochemical strain identifications* |
|---|---|---|---|---|
| ~ | 9.40 | 9.21 | 5.74 | |
| Mucosa | ||||
| 1 | 8.77 | 8.40 | 5.08 | Leuconostoc mesenteroides |
| 2 | 9.46 | 8.43 | 5.49 | Leuconostoc mesenteroides |
| 3 | 9.02 | 9.36 | 5.09 | Pediococcus pentosaceus |
| 4 | 9.06 | 7.23 | 4.83 | Lactobacillus plantarum |
| 5 | 8.28 | 7.67 | 5.06 | Lactobacillus plantarum |
| 6 | 8.78 | 8.47 | 4.01 | Lactobacillus salivarius |
| 7 | 8.71 | 8.49 | 6.43 | Leuconostoc mesenteroides |
| 8 | 9.36 | 8.39 | 6.47 | Lactobacillus crispatus |
| 9 | 9.39 | 9.11 | 5.44 | Leuconostoc mesenteroides |
| 10 | 9.37 | 8.68 | NG | Lactobacillus brevis |
| 11 | 9.36 | 8.84 | NG | Lactobacillus brevis |
| 12 | 8.82 | 8.48 | 3.33 | Leuconostoc mesenteroides |
| Milk | ||||
| 13 | 9.02 | 8.14 | NG | Lactobacillus brevis |
| 14 | 9.15 | 8.31 | NG | Lactobacillus brevis |
| Colostrum | ||||
| 15 | 9.32 | 9.14 | 5.34 | Lactococcus lactis |
| 16 | 9.32 | 8.95 | 4.52 | Leuconostoc mesenteroides |
| Mean | 9.07±0.33a | 8.50±0.54b | 5.09±0.89c |
* Identification done with API system (API 50CHL). Treatment ~ corresponds to positive control strain L. casei. Data are the mean of three replicates and correspond to log10 CFU/ml. NG: No growth.
a,b Different superscript letters in the mean value ± standard deviation indicate significant difference. (P<0.03).
Strain viability based on resistance to acid pH
Analysis of colony population resistance to different pH levels (Table 1), found growth at the control pH (6.5) to average 9.07 log10 CFU/ml. At pH 4.0 growth decreased (P<0.001) to 5.09 log10 CFU/ml. The two colonies from the milk samples did not grow at pH 4 and were not included in the final strains. Resistance to acid pH is relevant because to reach the action site and remain viable probiotic bacteria must withstand acid pH and the presence of BS in the duodenum24. Several authors have developed methodologies to evaluate probiotic strain resistance under gastrointestinal conditions7,25. The present results coincide with a study of L. plantarum and L. acidophilus in which these strains grew and remained viable at pH 5.0, but became inactive at pH 4.0 and 3.016. Strain sensitivity may be related to the acid tolerant response or acquired resistance. For example, in a study comparing a control of L. casei cells grown at pH 6.0 to acid adapted cells at pH 4.5 for 10 and 20 min, viability decreased up to 4.0 log10 CFU/ml at 10 min adaptation, and from 0.7 to 2.4 log10 CFU/ml at 20 min26. Cell adaptation to an acid environment caused changes in membrane lipid composition, with a dramatic increase in saturated and unsaturated fatty acids, as well as malolactic fermentation and intracellular histidine accumulation. The ability of probiotic bacteria to survive the stomach’s acid environment varies by strain27,28,29, which would explain the differences in resistance between LAB strains observed here at pH 4.0. Lactobacilli commonly grow better at pH 4.0 than at pH 3.030, and at pH 3.0 only four of 200 known LAB strains survive31.
Viability under bile salts exposure
When exposed to high BS concentrations, the LAB tested in the present study continued to grow at concentrations as high as 1.5 g. Lactic acid bacteria resistance to and growth under exposure to BS has been tested at concentrations from 0.1 to 4.0 %32,33; this is an important parameter for microorganisms in commercial products34,35, but one rarely tested. In another study36, L. plantarum resistance was exposed to four concentrations of porcine BS (0.01, 0.05, 0.10 and 0.15 g), and strain growth monitored for 24 h via OD measurements. The highest growth rate was observed at the lowest BS concentration, and, at the final density and 0.10 g BS, this strain’s growth rate was three times lower than in the control. This is higher growth inhibition at lower BS concentrations than observed in the present study: final colony OD was only 2.5 times lower at 0.3 g BS than in the control treatment. There are reports of resistance to BS at concentrations from 0.3 to 1% BS in LAB (Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus and Lactococcus lactis) and probiotic bacteria (L. acidophilus, L. casei, L. rhamnosus and Bifidobacterium)37,38. In these studies, S. thermophilus was the most sensitive LAB strain (growth inhibition at 0.5 g BS), L. lactis was the most resistant LAB (growth inhibition at 1 g BS), and all the probiotic strains exhibited resistance to 1.5 g BS. Resistance to BS exposure may differ between Lactobacillus species based on their ability to colonize and rapidly stabilize, as has been tested in the intestine of heifers7. For example, in an in vivo study in which L. acidophilus was administered to heifers, the total lactobacilli count in the jejunum increased from 13 to 39 %, but strains of L. plantarum and Lactococcus acidilactici exhibited better growth at pH 4.0 and 0.3 g BS5. In the present results, the LAB were more tolerant of BS exposure than of acid pH levels (4.0). However, their relatively good resistance to prolonged exposure to acid pH and very good resistance to high BS concentrations are effective indicators of their survival and colonization capacity during intestinal transit28,39.
Resistance to BS was also quantified by comparing strain average OD at two BS concentrations (0.3 and 1.5 g) (Table 2). Average OD in all the evaluated strains increased 3.1 times (P<0.05) after 24 h incubation at 0.3 g SB, compared to the initial reading, but when exposed to 1.5 g BS for 24 h, it increased 2.7 times. Optical density (OD) dropped significantly (P<0.023) as BS concentration increased, but it still increased (P<0.0001) from 1 to 24 h.
Table 2: Average optic density (OD) of growth in lactic bacteria strains at two bile salts (BS, %) concentrations at 1 and 24 hours’ incubation
| Treatment | Main Effects | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.3% BS | 1.5% BS | MSE | 0.3% BS | 1.5% BS | MSE | 1h | 24h | MSE | ||||||
| I | 1h | 24h | 1h | 24h | ||||||||||
| OD | 0.33 | 1.02 | 0.31 | 0.84 | 0.03 | 0.68 | 0.57 | 0.02 | 0.32 | 0.93 | 0.02 | |||
| P>F | 0.008 | 0.008 | 0.023 | 0.0001 | ||||||||||
MSE = Mean standard error.
Evaluations of the benefits of probiotic bacteria in heifers have been contradictory, perhaps due to a lack of diversity in probiotics for specific geographical regions and sale of unviable strains. This makes it difficult to find experimental sequences utilizing the same strain40. The marketing of probiotic products for heifers in Latin America is limited and dubious. Most suppliers offer only L. acidophilus strain KA1-A 8 (3 trillion CFU/dose) and L. casei (3 billion CFU/dose), without specifying their use in heifers. Others promote products containing Lactobacillus rhamnosus and Bifidobacterium lactis (1 x 1011 CFU/ dose or 50 units for direct application in 1,000 L milk). But they do not specify strain origin, leaving open the possibility that they have been isolated from other animal species or food, making them suboptimum options for use in heifers. For example, when human probiotic strains are administered to livestock the microorganisms cannot resist gastrointestinal conditions or colonize the intestine due to interspecies differences in physiology and food41,42. This is why the strains isolated in the present study were taken from the species in which they are intended for use and in the region they are to be applied; their probiotic potential can therefore be stated to be for Holstein cattle in the study area. Currently, the most widely used probiotic bacteria genera in livestock are Lactobacillus, Enterococcus, Bifidobacterium, Lactococcus and Leuconostoc43; Lactobacillus plantarum, L. acidophilus, L. casei, L. salivarius and Lactococcus lactis5,44,45; and L. fermentum VC3B-08, W. hellinica V1V-30 and L. farciminis B4F-0620.
Conclusions and implications
Sixteen lactic acid bacteria colonies were selected from the oral mucosa, milk and colostrum of Holstein cattle. Lactobacillus brevis isolated from samples of the oral mucosa and milk did not grow in acid pH (4.0). Based on their relative resistance to acid pH and good resistance to bile salts, five strains were selected: Leuconostoc mesenteroides, Pediococcus pentosaceus, Lactobacillus plantarum, Lactobacillus crispatus and Lactococcus lactis. These strains have broad probiotic potential in heifers and require further direct in vivo evaluation in the gastrointestinal tract.
Literatura citada:
1. Soto LP, Frizzo LS, Avataneo E, Zbrun MV, Bertozzi E, Sequeria G, Signorini ML, Rosmini MR. Design of macrocapsules to improve bacterial viability and supplementation with a probiotic for young calves. Anim Feed Sci Technol 2011;(165):176-183. [ Links ]
2. Shu Q, Gill HS. Immune protection mediated by the probiotic Lactobacillus rhamnosus HN001 (DR20TM) against Escherichia coli O157:H7 infection in mice. FEMS Inmunol Med Microbiol 2002;(34):59-64. [ Links ]
3. Frizzo LS, Zbrun MV, Soto LP, Signorini ML. Effects of probiotics on growth performance in young calves: A meta-analysis of randomized controlled trials. Anim Feed Sci Technol 2011;(169):147-156. [ Links ]
4. Rosmini MR, Sequeira GJ, Guerrero I, Marti LE, Dalla R, Frizzo L, Bonazza JC. Producción de prebióticos para animales de abasto: Importancia del uso de la microbiota intestinal indígena. Rev Mex Ing Quim 2004;(3):181-191. [ Links ]
5. Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Isolation of bovine intestinal Lactobacillus plantarum and Pediococcus acidilactici with inhibitory activity against Escherichia coli O157 and F5. J Appl Microbiol 2008;(106):393-401. [ Links ]
6. Seifzadeh S, Aghjehgheshlagh FM, Abdibenemar H, Seifdavati J, Navidshad B. The effects of a medical plant mix and probiotic on performance and health status of suckling Holstein calves. Italian J Anim Sci 2017;(16):44-51. [ Links ]
7. He ZX, Ferlisi B, Eckert E, Brown HE, Aguilar A, Steele MA. Supplementing a yeast probiotic to pre-weaning Holstein calves: feed intake, growth and fecal biomarkers of gut health. Anim Feed Sci Technol 2017;(226):81-87. [ Links ]
8. Wannaprasat W, Koowatananukul C, Ekkapobyotin C, Chuanchuen R. Quality analysis of commercial probiotic products for food animals. Southeast Asian J Trop Med Public Health 2009;(40):1103-1112. [ Links ]
9. Municipio de Texcoco. https://es.wikipedia.org/wiki/Municipio_de_Texcoco. [ Links ]
10. NOM-109-SSA1-1994: Proyecto de Norma Oficial Mexicana, Bienes y Servicios. Procedimiento para la toma, manejo y transporte de muestras de alimentos para su análisis microbiológico. 1994. Diario Oficial de la Federación. 1994. [ Links ]
11. Coeuret V, Dubernet S, Bernardeau M, Gueguen M, Vemoux JP. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Dairy Sci Technol Le Lait 2003;(83):269-306. [ Links ]
12. Pérez MJ, Vázquez JR, Rodríguez MC, Miranda RE, Romo AL, Nader GE. Procedimientos de laboratorio para bacteriología y micología veterinarias. Universidad Nacional Autónoma de México, México, DF. 1987. [ Links ]
13. Kandler O, Weiss N. nonsporing Gram positive rods. Bergey´s Manual of systematic bacteriology.10th ed (ed. P. H. A. Sneath, N. S. Mair, M. E. Sharpe and J. G. Holt Sneath), Baltimore, USA: The Williams and Wilkins Co; 1992. [ Links ]
14. Haro M, Ruiz V, Guerra F. Manual para la identificación de microorganismos de interés veterinario. México: Trillas; 2012. [ Links ]
15. Bove P, Gallone A, Russo P, Capozzi V, Albenzio M, Spano G, Fiocco D. Probiotic features of Lactobacillus plantarum mutant strains. Appl Microbiol Biotechnol 2012;(96):431-441. [ Links ]
16. Fernández de Palencia P, López P, Corbí AL, Peláez C, Requena T. Probiotic strains: survival under simulated gastrointestinal conditions, in vitro adhesion to Caco-2 cells and effect on cytokine secretion. Eur Food Res Technol 2008;(227):1475-1484. [ Links ]
17. Pereira DI, McCartney AL, Gibson GR. An in vitro study of the probiotic potential of a bile-salt-hydrolyzing Lactobacillus fermentum strain, and determination of Its cholesterol-lowering properties. Appl Environ Microbiol 2003;(8):4743-4752. [ Links ]
18. Tinrat D, Saraya S, Chomnawang MT. Isolation and characterization of Lactobacillus salivarius MTC 1026 as a potential probiotic. J Gen Appl Microbiol 2011;(57):365-378. [ Links ]
19. SPSS® Version 15 software SPSS (SPSS Inc., Chicago, IL). Copyright © 2006 de SPSS Inc. [ Links ]
20. Sandes S, Alvim L, Silva B, Acurcio L, Santos C, Campos M, Santos C, Nicoli J, Neumann E, Nunes A. Selection of new lactic acid bacteria strains bearing probiotic features from mucosal microbiota of healthy calves: Looking for immunobiotics through in vitro and in vivo approaches for immunoprophylaxis applications. Microbiol Res 2017;(200):1-13. [ Links ]
21. Ekinci F, Gurel M. Effect of using propionic acid bacteria as an adjunt culture in yogurt production. J Dairy Sci 2007;(91):892-899. [ Links ]
22. Jaramillo GD, Meléndez AP, Sánchez MO. Evaluación de la producción de bacteriocinas a partir de Lactobacillus y Bifidobacterias. Rev Venez Cienc Tecnol Aliment 2010;(1):193-209. [ Links ]
23. Marroki A, Zúñiga M, Kihal M, Pérez- Martínez G. Characterization of lactobacillus from algerian goat’s milk based on phenotypic, 16sr DNA sequencing and their technological properties. Brazilian J Microbiol 2011;(42):158-171. [ Links ]
24. Salminen S, Von Wright A, Morelli L, Marteau P, Brassart D, de Vos WM, et al. Demonstration of safety of probiotics-a review. Int J Food Microbiol 1998;(44):93-106. [ Links ]
25. Dunne C, O´Mahony L, Murphy L. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 2001;(73):386-392S. [ Links ]
26. Broadbent JR, Larsen RL, Deibel V, Steele JL. Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J Bacteriol 2010;(192):2445-2458. [ Links ]
27. Charteris WP, Kelly PM, Morelli L, Collins JK. Development and application of an in vivo methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol 1998;(84):759-76. [ Links ]
28. Chou LS, Weimer B. Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J Dairy Sci 1999;(82):23-31. [ Links ]
29. Xanthopoulos V, Litopoulou-Tzanetaki E, Tzanetakis N. Characterization of Lactobacillus isolates from infant faeces as dietary adjuncts. Food Microbiol 2000;(17):205-215. [ Links ]
30. Jin LZ, Ho YW, Abdullah N, Jalaludin S. Acid and bile tolerance of Lactobacillus isolated from chicken intestine. Lett Appl Microbiol 1998;(27):183-185. [ Links ]
31. Prasad J, Gill H, Smart J, Gopal PK. Selection and characterization of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J 1998;(8):993-1002. [ Links ]
32. Gómez-Zavaglia A, Kociubinski G, Perez P, De Antoni G. 1Isolation and characterization of Bifidobacterium strains for probiotic formulation. J Food Prot 1998;(61):865-873. [ Links ]
33. Kociubinski G, Pérez P., De Antoni G. Screening of bile resistance and bile precipitation in lactic acid bacteria and bifidobacteria. J Food Pro 1999;(62):905-912. [ Links ]
34. Carr F, Chill D, Maida N. The lactic acid bacteria: A literature survey. Crit Rev Microbiol 2002;(28):281-370. [ Links ]
35. Foditsch C, Pereira RVV, Ganda EK, Gómez MS, Marques EC, Santin T, Bicalho RC. Oral Administration of Faecalibacterium prausnitzii decreased the incidence of severe diarrhea and related mortality rate and increased weight gain in preweaned dairy heifers. PLOS ONE 2015;(10):e0145485. [ Links ]
36. Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebesem M. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J Bacteriol 2014;(186):5721-5729. [ Links ]
37. Naidu AS, Biblack WR, Clemens RA. Probiotic spectra of lactic acid bacteria. Critical Rev Food Sci Nutrition 1999;(38):13-126. [ Links ]
38. Lee YK, Nomoto K, Salminen S, Gorbach S. Handbook of probiotics. Lee YK editior. New York, USA: John Wiley & Sons. Inc; 1999. [ Links ]
39. Haller D, Colbus H, Ganzle MG, Scherenbacher P, Bode C, Hammes WP. Metabolic and functional properties of lactic acid bacteria in the gastro-intestinal ecosystem: a comparative in vitro study between bacteria of intestinal and fermented food origin. Syst Appl Microbiol 2001;(24):218-226. [ Links ]
40. Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, Tsakalidou E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J 2006;(16):189-199. [ Links ]
41. Ewaschuk JB, Naylor JM, Chirino-Trejo M, Zello GA. Lactobacillus rhamnosus strain GG is a potential probiotic for calves. Can J Vet Res 2004;(68):249-253. [ Links ]
42. Ewaschuk JB, Zello GA, Naylor JM. Lactobacillus GG does not affect D-lactic acidosis in diarrheic calves, in a clinical setting. J Vet Int Med 2006;(20):614-619. [ Links ]
43. Gaggia F, Mattarelli P, Biavati B. Probiotics and prebiotics in animal feeding for safe food production. Int J Food Microbiol 2010;(141):S15-S28. [ Links ]
44. Cebeci A, Gürakan C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol 2003;(20):511-518. [ Links ]
45. Vinderola CG, Reinheimer JA. Lactic acid starter and probiotic bacteria: a comparative in vitro study of probiotic characteristics and biological barrier resistance. Food Res Int 2003;(36):895-904. [ Links ]
Received: June 01, 2017; Accepted: February 27, 2018











 text in
text in