Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias agrícolas
Print version ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.12 n.7 Texcoco Sep./Nov. 2021 Epub Mar 22, 2022
https://doi.org/10.29312/remexca.v12i7.2677
Articles
Molecular characterization of caimito in the state of Morelos
1Posgrado en Ciencia Agropecuarias y Desarrollo Rural-Facultad de Ciencias Agropecuaria-Universidad Autónoma del Estado de Morelos. Avenida Universidad 1001, Colonia Chamilpa, Cuernavaca, Morelos, México. CP. 62209. Tel. 777 3702946.
2Escuela de Estudios Superiores de Xalostoc-Universidad Autónoma del Estado de Morelos. Av. Nicolás Bravo s/n, Parque Industrial Cuautla, Xalostoc, Ayala, Morelos, México. CP. 62717. Tel. 777 3297981, 777 329-7988.
The fruit growing of Mexico has 63 commercial species and 220 species with food potential. In the state of Morelos, 1.7 ha of caimito have been reported in the municipalities of Coatlán del Río and Tetecala de la Reforma. Caimito is an alternative source for the diversification of agriculture, as well as for fruit export. The knowledge of the genetic variability of the species will allow the development of programs oriented to the conservation of germplasm in situ or ex situ, certification programs, as well as provide producers with a wide selection of plants. The objective of the study was to molecularly characterize previously selected caimito trees in Coatlán del Río and Tetecala de la Reforma, Morelos, Mexico, using the RAPDs technique. Thirteen caimito trees were selected considering the morphological and organoleptic characteristics for the selection. Genomic DNA isolation was performed, and 30 RAPD primers were used. Molecular data were processed using the numerical taxonomy and multivariate analysis system (NTSYSpc 2.1). The methodology used allowed the characterization of nine trees. The cluster analysis obtained shows that at a level of 0.71 of genetic distance, four groups form. Trees 14 and 15 do not share genetic similarity with the other trees and these were in turn the ones that showed the greatest number of marker bands and were also some of those that stood out for their flavor.
Keywords: characterization of fruit trees; DNA extraction; molecular markers; RAPDs
La fruticultura de México cuenta con 63 especies comerciales y 220 especies con potencial alimenticio. En el estado de Morelos se han reportado 1.7 ha de caimito en los municipios de Coatlán del Río y Tetecala de la Reforma. El caimito es una fuente alternativa para la diversificación de la agricultura, así como para la exportación frutícola. El conocimiento de la variabilidad genética de las especies permitirá el desarrollo de programas orientados a la conservación de germoplasma de forma in situ o ex situ, programas de certificación, así como proporcionar a los productores una amplia selección de plantas. El objetivo de la investigación fue caracterizar molecularmente árboles de caimito seleccionados previamente en Coatlán del Río y Tetecala de la Reforma, Morelos, México, utilizando la técnica de RAPDs. Se seleccionaron 13 árboles de caimito tomando en cuenta para la selección con base en las características morfológicas y organolépticas. Se realizó el aislamiento de ADN genómico y se utilizaron 30 iniciadores RAPD. Los datos moleculares se procesaron utilizando el sistema de análisis multivariado y taxonómico numérico (NTSYSpc 2.1). La metodología utilizada permitió caracterizar nueve árboles. El análisis del conglomerado obtenido muestra que a un nivel de 0.71 de distancia genética se forman cuatro agrupaciones. Los árboles 14 y 15 no comparten similitud genética con los otros árboles y estos fueron a su vez los que mostraron mayor número de bandas marcadoras y así como también fueron unos de los que destacaron por sabor.
Palabras clave: caracterización de árboles frutales; extracción de ADN; marcadores moleculares; RAPDs
Introduction
Fruit growing in Mexico has 63 commercial species and 220 species with food potential, some species are not yet reflected in the statistics as they are grown locally in home gardens or are collected (Borys and Borys, 2001), among which are those of the family Sapotaceae; of this, caimito (C. cainito L.) is native to Central America and the Caribbean Islands and the Antilles. In the state of Morelos, 1.7 ha of caimito have been reported in the municipalities of Coatlán del Río and Tetecala de la Reforma (Álvarez-Vargas et al., 2006; SIACON, 2018).
Chrysophyllum L. (Sapotaceae) species are propagated widely as ornamental plants due to their colorful foliage density and edible fruits (El-Harwary et al., 2019). They are fleshy fruits, globose or rounded berry type, the pulp is fleshy, it stands out for its exquisite flavor and nutritional value, since the fruits are rich in carbohydrates, calcium and phosphorus, they can be consumed as fresh fruit, in juices or salads, they are also used for ornamental purposes for their appearance and golden color of the abaxial of the leaves (Morton, 1987; Pennington, 1990; Rojas-Rodríguez and Torres-Córdoba, 2012).
It is for the above mentioned that Azurdia et al. (1995) suggests the importance of caimito as an alternative source for the diversification of agriculture as well as for fruit exports. There is little knowledge about the techniques of sexual and asexual propagation of this fruit tree, as well as about its cultivation in general, with conditions and practices that ensure a good production of high-quality fruits. To achieve this, it is necessary to produce suitable rootstocks in order to perpetuate the selected varieties or types of this plant (Álvarez et al., 2004).
In this regard, Crane and Balerdi (2019) mention that the caimito can be propagated by seed, cuttings and grafting, trees that originate from seed bear fruit in 5 to 10 years, while vegetatively propagated trees can begin fruit production in 1 to 2 years, and in grafting, it has been reported that caimito produces slow-growing dwarf trees. Asexual propagation limits the number of generations that have passed since their first cultivation by humans and the degree of evolution that has occurred under human selection in these taxa (Zohary and Spiegel-Roy, 1975; Clement, 1989). When the caimito is propagated by seed, it generates a wide genetic diversity, so it is advisable to multiply the trees selected for their productive capacity and quality of their fruits through grafting (Avilan et al., 1992).
If different cultivated varieties are selected by contrasting phenotypes, the phenotypic variance will be greater in cultivated plants (Parker et al., 2010). The knowledge of the genetic variability of the species will allow the development of programs oriented to the conservation of germplasm in situ or ex situ, certification programs, as well as provide producers with a wide selection of plants (Azurdia et al., 1997; Carrara, 2004; Nascimento et al., 2008). There are different methods to characterize, identify plant species, evaluate their diversity and phylogeny, those based on morphology are the most used (Bayuelo-Jiménez et al., 2006).
The first characterization studies of sapotaceous used morphological, phenological, and agronomic characters; however, classification based on these characters can be confusing because they are affected by environmental factors. The use of isoenzymes may be more useful; in this regard, they were applied to study in situ 246 types of M. sapota and 287 of C. cainito in Nicaragua (Benavides, 1998; Weaver, 1993) by morphological characterization, registered a new species of caimito called Micropholis chrysophylloides in Puerto Rico; however, these can be affected by the environment and the phenological stage of the plant.
A large number of methods to identify and analyze molecular markers have also been generated, among which those based on the polymerase chain reaction (PCR) stand out, which allow amplifying specific or random sequences of DNA and require small initial amounts of it (Orona et al., 2006), they are not affected by the environment and can be analyzed at any stage of development of the crop, RAPD markers are simple, efficient, economical, fast and efficiently discriminate between genotypes (Yasmin et al., 2006).
Markers such as cytogenetic and molecular allow analyzing the differences between chromosomes, proteins or DNA (Avise, 2004). Another technique used is the sequencing of the ndhF chloroplast gene to study the phylogeny in the family (Anderberg and Swensony, 2003). Simple sequence repeats (SSRs) were used for the analysis of the variability of the chloroplast DNA in Manilkara hubery (Rennó et al., 2008). RAPDs were used to determine the effect of environmental conditions on the genotype of Manilkara sapota (Heaton et al., 1999) and to determine the spatial genetic structure of Chrysophyllum sanguinolentum (Degen et al.,2001), as well as to study the genetic diversity of Vitellaria paradoxa (Fontaine et al., 2004).
In Pouteria sapota, AFLPs have also been used to study the genetic variation of cultivated selections (Carrara, 2004). Majourhat et al. (2008) used RAPDs and SSRs to characterize morphotypes of Argania spinosa and observed that RAPDs had high polymorphism and more information than SSRs. There are currently many germplasm collections with genotypes of high agronomic value that could be used as parents in genetic improvement programs; however, in many cases their degree of diversity and the relationship between materials are unknown, which makes it difficult to use them (Becerra and Paredes, 2000).
Molecular markers are widely used to implement crop breeding and management programs (Xin-hua et al., 2007). This method is beneficial for ecologists and conservationists by allowing the identification of samples (Hebert and Gregory, 2005). González-Hernández et al. (2012) mention that, although the sapodilla (Sapotaceae) is an economically important species in Mexico, there is no information that allows generating appropriate programs for its improvement and crop management.
Due to the limited information about caimito in terms of molecular characterization that allows identifying the trees with better characteristics that could be used to conserve them and eventually integrate them into a program of long-term genetic improvement or vegetative propagation that allows having caimito orchards established in a homogeneous way and with quality fruits, the objective of this research was to molecularly characterize previously selected caimito trees in Coatlán del Río and Tetecala de la Reforma, Morelos, Mexico, using the RAPDs technique, in order to determine if there is a genetic differentiation of trees that allows the identification and selection of outstanding germplasm; through molecular markers.
Materials and methods
Thirteen caimito trees were selected based on morphological and organoleptic characteristics such as fruit flavor, higher yield, size and quality, through interviews with the producers conducted by Álvarez-Vargas et al. (2006) in Coatlán del Río and Tetecala de la Reforma, Morelos, Mexico. The characteristics for which they were selected are because the caimito fruits are red, opaque and dark, there is great variation in the hue and chromaticity of the fruits, probably due to the wide variety in colors and their intensity. The dimensions of the fruit, such as length and diameter, show a high variation with ranges of 60.3 and 54.1 cm, respectively.
Sixty percent of the total weight of the fruit corresponds to the peel, 3% to the seed and 37% to the pulp, the peel of the fruit is composed of a thin and hard epicarp, as well as part of the mesocarp that at commercial maturity forms a resistant peel, the other part of the mesocarp is purple or white, aqueous and sweet and represents the pulp of the fruit. The great variation in these characters makes it possible to search for materials with a higher proportion of pulp, to increase their potential use as fresh or processed fruit. The caimito can have up to eight seeds, as well as seedless fruits.
Caimito fruits have great variation in chemical characteristics such as soluble solids, total sugars. The average values of soluble solids are 10.1 Brix and indicate that the fruits provide a good amount of energy to those who consume them, some fruits reach values of 185.9 mg g-1 of total sugars. Currently, few quality studies have been carried out to determine the characteristics of the fruit that are attractive to the consumer; however, large fruits with few seeds and a high concentration of total sugars are preferred (Álvarez-Vargas et al., 2006). The trees were 1, 2, 3, 4, 5, 9, 10, 13, 14, 15, 17,18 and 19.
Leaf collection
Samples of young, healthy leaves were collected from 13 previously selected caimito trees. The leaves were placed in plastic bags, marked with the code of each tree and transported at ambient temperature to the laboratory. The leaves were washed, and the water was removed from the surface, then 150 mg of leaf from each tree was weighed and frozen at -20 °C until DNA extraction.
Genomic DNA isolation
Samples of 150 mg of leaves from each of the 13 caimito trees were used to extract DNA according to the methodology reported by Andrade-Rodríguez et al. (2005), replacing isoamyl alcohol with octanol. In the last step of the extraction methodology, the DNA obtained was dissolved in 25 (l of 0.1 TE (1 mM Tris-HCl pH 8, 0.1 mM EDTA) with 20 ng (l-1 of RNase, heated to 37 °C for 40 min. And stored at -20 °C until use.
The DNA integrity of all trees was estimated by 1% agarose gel electrophoresis (ultrapure GIBCO), for which 1 (l of DNA sample was used. Electrophoresis was performed at ambient temperature in TAE buffer (0.04 M tris-base, glacial acetic acid, 1 mM EDTA) at 75 volts for two hours. The gel was stained with ethidium bromide (1 (g ml-1). DNA samples were visualized and documented with a gel analyzer (Syngene GVM20).
The concentration of the DNA and its purity were quantified in a Genesys 6® spectrophotometer of ultraviolet light, for which a dilution of 1:150 [5 (l of DNA from the sample plus 745 (l of TE 10X (10 mM Tris HCl pH 8, 1 mM EDTA pH 8)] was prepared. The concentration was determined by the formula: [DNA (ng (l-1)]= (OD260) (DF) (50 (g∙(l-1). Where: OD260= optical density of the DNA solution read at the wavelength of 260 nm. DF= dilution factor. 50 g( (l-1= DNA concentration determined for a value of 1 at 260 nm.
Purity was assessed as the ratio of readings at wavelengths of 260 and 280 nm (260/280). Values between 1.8 and 2 of optical density (OD) indicate a high degree of purity. Values less than 1.8 indicate contamination of the DNA sample with proteins and other UV light-absorbing elements. While values greater than 2.0 indicate contamination by chloroform, phenol or other organic substance. Once the reading for each DNA sample was obtained, the working solutions were prepared at 20 ng (l1.
RAPD Analysis
Thirty primers were used, 20 with the sequences of kit B, A-07, A-08 (Operon Technologies Inc.) and eight with the sequences designed by Rodríguez-Rojas et al. (2012) for studies with sapotaceous, all with an arbitrary sequence of 10 nucleotides, of these primers, 12 that presented the largest number of brightest bands were selected (Table 1).
Table 1 Sequence of eight of the primers used in the molecular characterization of caimito trees, designed by Rodríguez-Rojas et al. (2012).
| Primer | Code | Sequence of bases | Primer | Code | Sequence of bases |
| 23 | SAP-01 | 5’ ATG CGA ACC G 3’ | 27 | SAP-05 | 5’ TAT AGG CCC T 3’ |
| 24 | SAP-02 | 5’ GAC ACA TCG G 3’ | 28 | SAP-06 | 5’ CCT ACT CCA G 3’ |
| 25 | SAP-03 | 5’ TGG GAC CTC C 3’ | 29 | SAP-07 | 5’ TGG GAA TCC C 3’ |
| 26 | SAP-04 | 5’ GGA GCT ACC T 3’ | 30 | SAP-08 | 5’ GCC CCT ACT A 3’ |
The reaction mixture consisted of 10 (l of dNTPs (5 (M of each dNTP), 2.5 (l PCR buffer (10X), 1.5 (l of Mg Cl2 (75 mM), 2 L of primer (20 pmol), 0.3 (l native DNA polymerase INVITROGEN (1.5 U), 4 (l of DNA (80 ng) adjusting to a volume of 25 (l with 4.7 L of sterile deionized distilled water. DNA amplification was performed by polymerase chain reaction (PCR) in a Techne TC-412 thermocycler. The thermocycler program consisted of a DNA pre-denaturation cycle at 94 °C for 4 min, plus 35 cycles consisting of the following stages: 1 min at 94°C to separate DNA helices, 1 min at 36 °C for primer alignment, 2 min at 72 °C for DNA polymerization and a final extension cycle of 10 min at 72 °C.
The separation of amplified products was performed by ultrapure agarose gel electrophoresis (INVITROGEN) at 1.5% (w/v). Electrophoresis was performed with TAE 1X buffer, applying 75 volts for 4.5 h. The gels were stained with ethidium bromide (1 (g L-1) to evidence the DNA bands, and subsequently photographed with the Syngene GVM20 photodocumenter. The size of the DNA fragments, produced by the RAPDs, was obtained using the Gen tools program version 3.06 of Syngene.
The analysis of the molecular data for the characterization of the 13 caimito selections was performed by comparing DNA band patterns produced by the 12 RAPD primers using the Gene tools Labworks 4® program. The presence of RAPD markers was determined by analyzing the electrophoretic profiles generated by each of the caimito genotypes. For the construction of the dendrogram, the data were analyzed based on the presence (indicated by 1) or the absence (indicated by 0) of fragments amplified with the decamer primers used, the data were processed using the numerical taxonomy and multivariate analysis system (NTSYSpc 2.1). Cluster analyses were performed on the relationship of matrices with the unweighted pair group method with arithmetic mean, UPGMA (Avise, 1994).
Results and discussion
The selected primers were OPB (01, 04, 05, 06, 07, 08, 09, 10, 11 and 12), SAP-01 and SAP-04, for presenting the largest number of amplified products, with brighter and better-defined bands. These primers generated 190 RAPD fragments ranging from 254 to 3 407 bps. Ninety-seven percent of the fragments were polymorphic and allowed characterizing 9 of the 13 trees studied (1, 3, 5, 10, 13, 14, 15, 17 and 19) with 71 marker fragments. In trees 2, 4, 9 and 18, no marker fragments that would allow their characterization with the primers used were obtained (Table 2). The primers that generated the largest number of markers were OPB-06 and OPB-10, the latter allowed identifying a greater number of trees (6) along with SAP-01 and SAP-04 (Figure 1). In contrast, primer OPB-11 produced only threes fragments which characterized two trees. No single primer allowed to characterize all plants.
Table 2 Amplified fragments and molecular characterization by RAPDs of caimito trees in Morelos, Mexico.
| Primer | Amplified fragments | Polymorphic fragments | Polymorphism (%) | Markers (pb) | Characterized plant |
| OPB 01 GTTTCGCTCC | 17 | 17 | 100 | p:1906 p:1813 p:877 a:1615 a:1044 | 13 1 13 13 13 |
| OPB 04 GGA CTG GAGT | 15 | 14 | 93 | p:1319 p: 490 | 5 19 |
| OPB 05 TGCGCCCTTC | 13 | 13 | 100 | p:1339,1081,866 p:492 a: 1265 | 14 10 14 |
| OPB 06 TGCTCTGCCC | 17 | 16 | 94 | p:1877,1628,1324 1063, 909,519 p:622,548, 495 a:1131,995 | 15 10 15 |
| OPB O7 GGTGACGCAG | 16 | 16 | 100 | p:2140,1396,918,496 a:1727 | 14 13 |
| OPB 08 GTCCACACGG | 16 | 16 | 100 | p:1493,1100,969,254 a:732 | 15 15 |
| OPB 09 TGGGGGACTC | 13 | 13 | 100 | p:1561,1403 p:1357,1026,626 a:1940 | 15 17 15 |
| OPB 10 CTGCTGGGAC | 21 | 21 | 100 | p:2726,2446,2280 1934,1762,1628, 1258, 899, 716,654 p:1208 a:1114 a:467 | 14 17 14 13 |
| OPB 11 GTAGACCCGT | 12 | 12 | 100 | p: 1187 p:987,831 | 10 15 |
| OPB 12 CCTTGACGCA | 12 | 11 | 91 | p:1192,693, 596 a:1028,665 | 14 14 |
| SAP 01 ATGCGAACCG | 21 | 20 | 95 | p:669 p:588 p:1884,1689,1406, 960 | 3 10 14 |
| SAP 04 GGAGCTACCT | 17 | 16 | 94 | p:310 p:687,306 p:1147 a:599 | 13 14 15 13 |
Pb= pair base; a= absence; p= presence.
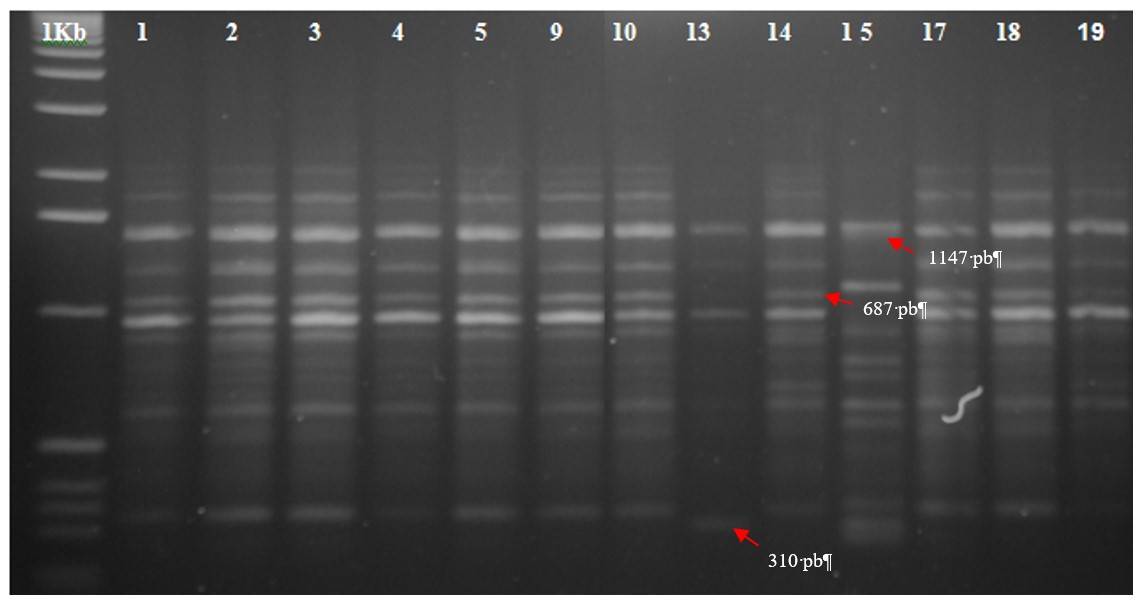
Figure 1 Banding pattern generated by primer SAP-04 for 13 caimito trees in the state of Morelos, Mexico. 1kb: molecular weight marker 1 kb DNA Ladder.
The methodology used allowed characterizing nine trees, the 14 was the one that showed the greatest presence of markers (30 fragments) that were produced with six primers. This indicates that the DNA of this tree had a greater number of particular DNA sequences that were complementary to primers OPB (05, 07, 10, 12), SAP-01 and SAP-04. Another tree that showed a greater number of marker bands was tree 15, with 15 marker fragments, generated by oligonucleotides OPB (06, 08, 09 and 11).
In contrast, trees 2, 4, 9 and 18 did not produce any markers, which means that the DNA molecule of these plants did not present particular DNA sequences that identified them with the use of the primers used. The organoleptic characteristics of the fruits are not reported; however, the tasting of the fruits was carried out, in flavor, the fruits of trees 3, 4, 10, 14 and 15 stood out; however, there were no markers for all these trees (Table 3).
Table 3 Fragments of molecular markers for 13 caimito trees from the state of Morelos, Mexico.
| Tree | Marker fragments |
| 1 | OPB 01 (1813) |
| 2 | ____________ |
| 3 | SAP 01 (669) |
| 4 | ___________ |
| 5 | OPB 04 (1319) |
| 9 | ___________ |
| 10 | OPB 05 (492), OPB 06 (622, 548, 495), OPB 11 (1187), SAP 01 (588) |
| 13 | OPB 01 (1906, 877, 1615, 1044), OPB 07 (1727), OPB 10 (467), SAP 04 (310, 599) |
| 14 | OPB 05 (1339, 1081, 866, 1265), OPB 07 (2140, 1396, 918, 496), OPB 10 (2726, 2446, 2280, 1934, 1762, 1628, 1258, 899, 716, 654, 1114), OPB 12 (1192, 693, 596, 1028, 665), SAP 01 (1884, 1689, 1406, 960), SAP 04 (687) (306) |
| 15 | OPB 06 (1877, 1628, 1324, 1063, 909, 519), OPB 06 (1131, 995), OPB 08 (1493, 1100, 969, 254, 732), OPB 09 (1561, 1403, 1940), OPB 11 (987, 831) |
| 17 | OPB 09 (1357, 1026, 626), OPB 10 (1208) |
| 18 | __________ |
| 19 | OPB 04 (490) |
The size of the fragments amplified by the primers corresponding to base pairs (pb) is indicated in parentheses.
The RAPD methodology allowed the characterization of 72.9%of the caimito trees studied, this because they amplify in a wide region of the genome; however, the possibility of finding molecular markers for all trees could have been higher if more primers had been used, in this regard, (Vinicius et al., 2014), when conducting a study to find what the best marker for the identification of Sapotaceae species was, mention and conclude the importance of molecular markers, which they consider essential to adopt conservation strategies and an appropriate management plan for a given species, as they are highly discriminatory and useful for the molecular identification of the Sapotaceae specie.
The RAPDs in the present research presented a high discriminative percentage, which allowed detecting the genetic diversity for the selection of outstanding trees, promising to selection, management and conservation programs that could be efficiently implemented, molecular markers serve as a parameter to classify and allow identifying the plant (mother) material of interest to propagate.
The total and polymorphic RAPD fragments obtained in the characterization of the caimito trees were greater than those obtained by Fontaine et al. (2004), who observed a total of 67 polymorphic and 15 monomorphic RAPD loci ranging from 1 670 to 280 bp, in 179 individuals of the sapotaceous Vitellaria paradoxa, with the primers OPB-07, OPB-11, OPN-15, OPR-15, OPW-9, OPW-12, OPW-13, OPX-3, OPX-6, OPX-11, OPY-6, OPY-13, OPY-20, OPW-5, OPW-19. In this research, the number of bands ranged from 12 to 21, while Fontaine et al. (2004) only identified 1 to 6 bands, which is attributed to the difference in species.
As for the number of marker fragments, in this research, 58 different ones were obtained with the use of 12 primers, while Heaton et al. (1999), when studying sapodilla using RAPDs with 80 primers, only obtained 28 different bands, this indicates the usefulness of the oligos used in this research. In this research, 185 polymorphic fragments were obtained, while Degen et al. (2001), using the same methodology to study 68 individuals of Chrysophyllum sanguinolentum with 11 primers (OPE-02, OPE-05, OPE-07, OPY-04, OPY-06, OPY-07, OPY-10, OPY-13, OPY-14, OPY-15, OPY-16), obtained only 48 polymorphic fragments. From the above described, it can be said that the number of amplified fragments varies according to the species to be studied, as well as the sequence of indicators.
The cluster analysis obtained (Figure 2) shows that at a level of 0.71 of genetic distance, four groups form: 1) trees 1, 4, 13, 18 and 19; 2) trees 2, 5, 3, 10, 9 and 17; and 3) tree 15 and 4) tree 14. The group that integrated more trees was the 2, the greatest genetic similarity between trees also occurred in this group. Plants two and five had the lowest genetic distance (0.92), in contrast, trees 14 and 15 were the most distant from the other trees, at the same time they were the ones that showed the highest number of marker bands and they also stood out for flavor.
Conclusions
The information generated on the molecular characterization of the caimito is one of the first references of DNA characteristics, for trees with outstanding fruit characters, which can be useful to start the process of selection and registration of varieties of this fruit species in Mexico. The RAPDs were useful for the characterization of nine caimito trees, achieving the molecular characterization of more than 72.9% of the plants studied. Trees 14 and 15 showed the highest number of marker bands and share the least genetic similarity with the other trees.
The species with potential for the conservation of germplasm in situ or ex situ are trees 15 and 15, which can enter a conservation and certification program to provide other producers with plants with the outstanding characteristics for the cultivation of caimito.
Acknowledgements
The support of PROMEP through the project UAEMOR-CA-74 is appreciated. Morphological and molecular characterization of selections of three species of sapotaceous in the state of Morelos and the SNI (exp. 34643). To the caimito producers from the state of Morelos for allowing access to their orchards and sharing experiences, time and meetings.
REFERENCES
Álvarez-Vargas, J. E.; Alia-Tejacal, I.; López-Martínez, V.; Acosta-Durán, C. M.; Andrade-Rodríguez, M.; Colinas-León, M. T.; Delgado-Escobar, I. y Villegas-Torres, O. 2006. Caracterización de frutos de caimito (Chrysophyllum cainito L.), en el estado de Morelos. Rev. Chapingo Ser. Hortic. 12(2):217-221. https://doi.org/10.5154/r.rchsh.2006.02.019. [ Links ]
Alvarez, B. R. J.; Graterol, C.; Quintero, I.; Zambrano, J.; Materano, W. y Maffei, M. 2004. Evaluación de algunos métodos y prácticas de propagación en caimito (Chrysophyllum cainito L). I. Sexual. Rev. Facult. Agron. Luz. 21(4):47-53. [ Links ]
Anderberg, A. and Swensony, A. 2003. Evolutionary lineages in sapotaceae (Ericales): a cladistic analysis based on ndhF sequence data. Inter. J. Plant Sci. 164(21):763-773. [ Links ]
Andrade-Rodríguez, M.; Villegas-Monter, Á.; Gutiérrez-Espinosa, M. A.; Carrillo-Castañeda, G. and García-Velázquez, A. 2005. Polyembryony and rapd markers for identification of zygotic and nucellar seedling in citrus. Agrociencia. 39(4):371-383. [ Links ]
Avilán, L.; Leal, F. y Batista, D. 1992. Manual de fruticultuta: principios y manejo de la producción. Segunda edición. Editorial América, Caracas. 1387-1401 pp. [ Links ]
Avise, J. C. 1994. Molecular markers, natural history and evolution. Editorial Chapman and Hall. New York, 510 p. [ Links ]
Avise, J. C. 2004. Molecular markers, natural history and evolution . Sinauer Associates. Editorial C.and Hall. Sunderland, Massachusetts. 541 p. [ Links ]
Azurdia, C.; Martínez, E. y Ayala, H. 1995. Algunas sapotáceas de petén, Guatemala. Proceedings of the Interamerican Society for Tropical Horticulture. 13(1): 33-45. [ Links ]
Azurdia, C.; Mejia, L. y Nufio, B. 1997. Variabilidad en frutales tropicales nativos de pouteria (Sapotaceae) utilizando marcadores isoenzimáticos. Ciencia y Tecnología. 2(1):3-25. [ Links ]
Bayuelo-Jiménez, J. S.; Lozano, R. J. C. y Ochoa, I. E. 2006. Caracterización morfológica de Byrsonima crassifolia (L.) kunth nativa de churumuco, Michoacán, México. Rev. Fitotec. Mex. 29(esp. 2):31-36. [ Links ]
Becerra, V. V. y Paredes, C. M. 2000. Uso de marcadores bioquímicos y moleculares en estudios de diversidad genética. Agric. Téc. 60(3):270-281. https://doi.org/10.4067/s0365-28072000000300007. [ Links ]
Benavides, G. A. 1998. Prospección y caracterización preliminar in situ de cinco especies de sapotaceas en Nicaragua. Repositorio Institucional La Calera,. Universidad Nacional Agraria. 12-21 pp. [ Links ]
Borys, M. W. y Borys, H. L. 2001. El potencial genético frutícola de la República Mexicana. Editorial Fundación Salvador Sánchez Colín, Cictamex, SC. Coatepec Harinas, México. 48 p. [ Links ]
Carrara, S. 2004. Genetic variation among cultivated selections of mamey Sapote (Pouteria spp. [Sapotaceae]). Proc. Fla. State Hort. Soc. 117:195-200. https://doi.org/10.25148/ etd.FI14052584. [ Links ]
Clement, C. R. 1989. A Center of crop genetic diversity in western amazonia. Bioscience. 39(9):624-631. [ Links ]
Crane, J. H. and Balerdi, C. F. 2019. Caimito (star apple) growing in the florida home planting a caimito tree in the home landscape. Institute of Food and Agricultural Sciences. 1-6 pp. [ Links ]
Degen, B.; Caron, H.; Bandou, E.; Maggia, L.; Chevallier, M. H.; Leveau, A. and Kremer, A. 2001. Fine-scale spatial genetic structure of eight tropical tree species as analysed by RAPDs. Heredity. 87(4):497-507. https://doi.org/10.1046/j.1365-2540.2001.00942.x. [ Links ]
El-Hawary, S. S. E.; El-Zalabani, S. M.; Selim, N. M.; Ibrahim, M. A.; Wahba, F. A.; El-Badawy, S. A.; Mahdy, N. E. S.; Yasri, A. and Sobeh, M. 2019. Phenolic constituents of Chrysophyllum oliviforme L. Leaf down-regulate TGF-β expression and ameliorate CCL4-induced liver fibrosis: evidence from in vivo and in silico studies. Antioxidants. 8(12):646. https://doi.org/10.3390/antiox8120646. [ Links ]
Fontaine, C.; Lovett, P. N.; Sanou, H.; Maley, J. and Bouvet, J. M. 2004. Genetic diversity of the shea tree (Vitellaria paradoxa C. F. Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity . 93(6):639-648. https://doi.org/10.1038/sj.hdy.6800591. [ Links ]
González-Hernández, D.; García-Pérez, E. y Guntin-Marey, P. 2012. Caracterización genética de Manilkara zapota de Veracruz, México, con marcadores SSR. Agrociencia . 46(7):663-675. [ Links ]
Heaton, H. J.; Whitkus, R. and Gómez-Pompa. 1999. Extreme ecological and phenotypic differences in the tropical tree chicozapote (Manilkara zapota (L.) P. Royen) are not matched by genetic divergence: a random amplified polymorphic DNA (RAPD) analysis. Mol. Ecol. 8:627-632. [ Links ]
Hebert, P. and Gregory, R. 2005. Thepromise of DNA barcoding for taxonomy. Systematic Biol. 54(5):852-859. [ Links ]
Majourhat, K.; Jabbar, Y.; Hafidi, A. and Martínez-Gómez, P. 2008. Molecular characterization and genetic relationships among most common identified morphotypes of critically endangered rare moroccan species argania spinosa (Sapotaceae) using RAPD and SSR markers. Annal. Forest Sci. 65(8):805). https://doi.org/10.1051/forest:2008069. [ Links ]
Morton, J. F. 1987. Sapote. In: fruits of warm climates. Julia F. Morton, Miami. 398-402 pp. [ Links ]
Nascimento, E. V.; Geraldo, M. A. B. y Hissayuki, H. R. 2008. Caracterização física e química de frutos de mamey. Rev. Bras. Frutic. 30(4):953-957. https://doi.org/10.1590/s0100-29452008000400019. [ Links ]
Orona, C. F.; Pecina, Q. V.; Rocha, P. M. A. y Cadena, H. M. A. 2006. Caracterización molecular de genotipos comerciales y elite de papa (Solanum tuberosum L.) en México. Agric. Téc.Méx. 32(2):171-180. [ Links ]
Parker, I. M.; López, I.; Petersen, J. J.; Anaya, A.; Cubilla-Ríos, L. and Potter, D. 2010. Domestication syndrome in Caimito (Chrysophyllum cainito L.): fruit and seed characteristics. Econ. Bot. 64(2):161-175. [ Links ]
Pennington, T. D. 1990. Flora neotropicana. Monografía 52. Jardín Botánico de Nueva York. 770 p. [ Links ]
Rennó, A. V. C.; Kanashiro, M.; Grattapaglia, D. y Yamaguishi, Y. A. 2008. Variabilidade no cpDNA em Manilkara huberi, espécie sob manejo sustentável na Amazônia brasileira. Pesqui. Agropecu. Bras. 43(7):859-867. https://doi.org/10.1590/s0100-204x2008000700010 . [ Links ]
Rodríguez-Rojas, T. J.; Andrade-Rodríguez, M.; Alia-Tejacal, I.; López-Martínez, V.; Espinosa-Zaragoza, S. y Esquinca-Avilés, H. 2012. Caracterización molecular de zapote mamey (Pouteria sapota (Jacq) Moore & Stearn) molecular characterization of zapote mamey (Pouteria sapota (Jacq) Moore & Stearn). Rev. Facul. Agron. Luz. 29(3):339-354. [ Links ]
Rojas-Rodríguez, F. y Torres-Córdoba, G. 2012. Caimito (Chrysophyllum cainito L.). Rev. Forest. Mesoam. Kurú. 9(23):45-46. [ Links ]
Vinicius, V. C.; Souza, M. R. C.; Alves-Araújo, A.; Marccus, A.; Mariano-Neto, E.; Den-Berg, C. and Amato, G. F. 2014. DNA barcoding in atlantic forest plants: what is the best marker for sapotaceae species identification? Gen. Mol. Biol. 37(4):662-670. https://doi.org/10.1590/S1415-47572014005000019. [ Links ]
Weaver, P. L. 1993. Micropholis chrysophylloides Pierra. Caimillo. Sapotaceae. Institute of Tropical Forestry, Southern Forest Experiment Station. Rio Piedras, Puerto Rico. 7 p. [ Links ]
Xin-hua, H.; Yong-ze, G.; Yang-mi, L. and S-jin, O. 2007. Assessment of the genetic relationship and diversity of mango and its relatives by cpISSR marker. Agric. Sci. 6(2):137-142. [ Links ]
Yasmin, S.; Shahidul, I.; Khondoker, N. and Samsul, A. 2006. Molecular characterization of potato germplasm by random amplified polymorphic DNA markers. Biotechnol. 5(1):27-31. https://doi.org/10.3923/biotech.2006.27.31. [ Links ]
Zohary, D. 1975. Unconscious selection and the evolution of domesticated plants. Econ. Bot. 58(1):5-10. https://doi.org/10.1663/0013-0001(2004)058[0005:USATEO]2.0.CO;2. [ Links ]
Received: April 01, 2021; Accepted: August 01, 2021











 text in
text in 



