Introduction
Calycolpus moritzianus is a species with a wide distribution in Norte de Santander, Colombia, where it is intensely used as a source of wood, live fences and fuel source, which has generated its indiscriminate logging and overexploitation, without considering the genetic diversity of the populations, which is harmful for its conservation (García & Suárez, 2008). Nevertheless, recently several studies have highlighted the potential of C. moritzianus essential oils (EOs) in the medical and cosmetic industry, an added value for this plant resource (Granados et al., 2012; Yáñez et al., 2011). The main components of C. moritzianus EOs are monoterpenes, including terpinen-4-ol, an essential oil with commercial viability as antimicrobial agent (Southwell et al., 1993; Yáñez et al., 2002, 2009). In 5 localities of Norte de Santander where C. moritzianus is present, 40 EOs are documented, being limonene their major component (20-40%; Granados et al., 2012; Yáñez et al., 2009), although their relative concentrations may vary according to the region of origin of the tree (Vanegas & Yáñez, 2011). Other EOs as α-Copaeno and α-Terpineol, show bactericidal and bacteriostatic effectiveness against Bacillus subtilis, Staphylococcus and others, and antiviral and antitumor activity in epithelial carcinoma cells from the human cervix have been observed (Mojica et al., 2011; Yáñez et al., 2009).
Calycolpus moritzianus (O. Berg) Burret, (syn. Psidium caudatum (McVaugh), commonly called “arrayán” or “cínaro”, is a species that belongs to the family Myrtaceae (Landrum & Kawasaki, 1997; Rivero, Pacheco et al., 2012). The known geographical distribution of this species includes populations that span from Ecuador, through Colombia to Venezuela (Hokche et al., 2008; Jorgensen & León, 1999; Tropicos, 2020). The largest myrtle populations are recorded in Colombia and Venezuela, in the latter one, the species is distributed in those states where a hills predominate such as Barinas, Lara, Mérida, Táchira and Trujillo (Hokche et al., 2008; Pérez, 2007; Rivero, Pacheco et al., 2012). In Colombia, C. moritzianus has been recorded in small forests populations in the departments of Antioquia (Cardona et al., 2011), Caldas (Durán et al., 2004), Boyacá, Cesar, Cundinamarca, Norte de Santander and Santander (Instituto de Ciencias Naturales, 2021). C. moritzianus is a wild tree species characterized morphologically by having upright leaves, solitary flowers or biparous cymes, with 4 or 5 sepals, with an approximate height of 10 to 15 m in mature individuals (McVaugh, 1956, 1968; Rivero, Pacheco et al., 2012). It is well adapted in a range of 700 to 2,200 m asl, with average temperatures between 15 °C to 22 °C.
Colombia is considered the second most diverse country in the world in species of plants with approximately 28,000 official records (Bernal et al., 2015; GBIF, 2020). Nevertheless, its plant biodiversity has not been widely explored, and knowledge of the genetic diversity of many native plants is unknown (FAO, 2013). For this reason, research on the use of these species is also limited. This issue offers an opportunity to promote projects for the sustainable and profitable use of this plant biodiversity in Latin America (Bravo et al., 2020; Castañeda et al., 2007; FAO, 2013). Thus, molecular characterization studies in native plants such as C. moritzianus is necessary to understand how to reproduce, propagate, and use plant species of Latin American biodiversity with potential cosmetic and medicinal use for sustainable commercial exploitation (Bravo et al., 2020; Castañeda-Cardona et al., 2018).
The study of the morphology and genetic variability of plants that can have medicinal and potential commercial purposes, allows for a better use of plant genetic resources (Castañeda-Cardona et al., 2018; Chaves-Bedoya et al., 2017; Palevitch, 1991), and offers important data to understand the ecological and evolutionary processes of natural populations of native trees (Jones & Hubbell, 2006; Mohammed et al., 2020; Staggemeier et al., 2010). Nevertheless, because morphological characters are highly influenced by the environment (phenotypic plasticity) and plant phenology (Arnold et al., 2019; Gianoli & Valladares, 2012), molecular characterizations and the use of DNA markers, which is environmentally independent, are an alternative approach to assess plant genetic variation (Alfalahi et al., 2019; Ansari et al., 2015). The genetic diversity of some Myrtaceae species in South America has been evaluated by using restriction fragment length polymorphisms (AFLP) in Acca sellowiana (Quezada et al., 2014), randomly amplified polymorphic DNA (RAPD) in Eugenia dysenterica (Trindade & Chaves, 2005), and Psidium guajava (Valera-Montero et al., 2016), inter simple sequence repeat (ISSR) in Myrcia lundiana (Alves et al., 2016) and Myrcia ovata (White et al., 2018), and the random amplified microsatellites (RAMs) in Psidium guajava (Sanabria et al., 2006). RAMs molecular markers, are a PCR-based method which uses primers containing microsatellite sequences and degenerate anchors at the 5’ end (Hantula et al., 1996; Zietkiewicz et al., 1994). RAMs markers are dominant (binary) and multi-allelic and they are an efficient tool to detect and estimate genetic variation in DNA among plants (Alfalahi et al., 2019; Muñoz et al., 2008; Posso et al., 2011; Zietkiewicz et al., 1994).
The previous studies in Myrtaceae indicate that these species present a high genetic diversity, however, there are no reports on the analysis of genetic diversity in C. moritzianus, in the known populations of this neotropical endemic species. The main objective of this study was to determine the genetic diversity of C. moritzianus in 5 geographic locations of north-west Colombia.
Materials and methods
Five municipalities were visited in Norte de Santander department in Colombia during the first trimester of 2011 (Ocaña, Chinácota, Toledo, Salazar y Pamplonita; Table 1). Three locations were surveyed per municipality and randomly collected 3 wild individuals per location with a minimum distance of 800 m between each other. Biological samples consisted of fresh and young leaves (reddish coloration) in an optimal phytosanitary state, which were preserved in silica gel until laboratory processing. Geographical location data was recorded in order to plot our sampling localities in a distribution map (Fig. 1).
Table 1 Description of individuals of C. moritzianus and localities sampled in Norte de Santander, Colombia.
| Sample code | Locality | Collection date | Altitude (m asl) | Geographic coordinates |
|---|---|---|---|---|
| 1 | Agua de la Virgen, Ocaña | 19/02/2011 | 1,589 | 8º12’47.3” N, 73º22’50.9” W |
| 2 | Agua de la Virgen, Ocaña | 19/02/2011 | 1,659 | 8º12’55.9” N, 73º23’5.7” W |
| 3 | Agua de la Virgen, Ocaña | 19/02/2011 | 1,624 | 8º13’3.2” N, 73º22’55.3” W |
| 4 | La Ermita, Ocaña | 20/02/2011 | 1,303 | 8º10’40” N, 73º19’6.2” W |
| 5 | La Ermita, Ocaña | 20/02/2011 | 1,390 | 8º10’39.6” N, 73º19’15.9” W |
| 6 | La Ermita, Ocaña | 20/02/2011 | 1,473 | 8º10’46.3” N, 73º19’29.2” W |
| 7 | Venadillo, Ocaña | 22/02/2011 | 1,227 | 8º15’24.7” N, 73º22’20.1” W |
| 8 | Venadillo, Ocaña | 22/02/2011 | 1,250 | 8º16’8.4” N, 73º23’24.5” W |
| 9 | Venadillo, Ocaña | 22/02/2011 | 1,347 | 8º15’59.8” N, 73º23’34.4” W |
| 10 | Iscalá Centro, Chinácota | 04/02/2011 | 1,854 | 7º32’4.8” N, 72º34’17.1” W |
| 11 | Iscalá Centro, Chinácota | 04/02/2011 | 1,829 | 7º32’11.9” N, 72º34’17.4” W |
| 12 | Iscalá Centro, Chinácota | 04/02/2011 | 1,816 | 7º32’26.1” N, 72º34’22.9” W |
| 13 | Manzanares, Chinácota | 04/02/2011 | 1,415 | 7º36’16.4” N, 72º35’14.6” W |
| 14 | Manzanares, Chinácota | 04/02/2011 | 1,421 | 7º36’1.3” N, 72º35’8.6” W |
| 15 | Manzanares, Chinácota | 04/02/2011 | 1,466 | 7º36’6.3” N, 72º35’9.8” W |
| 16 | Cineral, Chinácota | 05/02/2011 | 1,666 | 7º33’36.1” N, 72º34’35.5” W |
| 17 | Cineral, Chinácota | 05/02/2011 | 1,594 | 7º33’48.2” N, 72º34’31.3” W |
| 18 | Cineral, Chinácota | 05/02/2011 | 1,594 | 7º33’57.3” N, 72º34’48.6” W |
| 19 | Buenavista, Toledo | 24/01/2011 | 1,717 | 7º18’22.9” N, 72º28’34.1” W |
| 20 | Buenavista, Toledo | 24/01/2011 | 1,744 | 7º18’27.7” N, 72º28’31.2” W |
| 21 | Buenavista, Toledo | 24/01/2011 | 1,775 | 7º18’17.8” N, 72º28’28.3” W |
| 22 | Tapatá, Toledo | 24/01/2011 | 2,088 | 7º25’23.1” N, 72º31’45.6” W |
| 23 | Tapatá, Toledo | 24/01/2011 | 2,141 | 7º25’40.7” N, 72º31’43.2” W |
| 24 | Tapatá, Toledo | 24/01/2011 | 2,153 | 7º26’4.4” N, 72º31’46.8” W |
| 25 | Palmar Alto, Toledo | 24/01/2011 | 1,660 | 7º18’49.8” N, 72°27’14.9” W |
| 26 | Palmar Alto, Toledo | 24/01/2011 | 1,708 | 7º18’49.7” N, 72º27’6.12” W |
| 27 | Palmar Alto, Toledo | 24/01/2011 | 1,729 | 7º18’56.1” N, 72º27’5.9” W |
| 28 | Aguas Calientes, Salazar de las Palmas | 12/02/2011 | 998 | 7º46’12.4” N, 72º49’59.9” W |
| 29 | Aguas Calientes, Salazar de las Palmas | 12/02/2011 | 991 | 7º46’13.2” N, 72º50’0.2” W |
| 30 | Aguas Calientes, Salazar de las Palmas | 12/02/2011 | 1,117 | 7º45’35.6” N, 72º50’0.4” W |
| 31 | Angostura, Salazar de las Palmas | 13/02/2011 | 1,054 | 7º46’50” N, 72º48’56.2” W |
| 32 | Angostura, Salazar de las Palmas | 13/02/2011 | 1,106 | 7º46’54.3” N, 72º49’1.6” W |
| 33 | Angostura, Salazar de las Palmas | 13/02/2011 | 1,201 | 7º47’10.7” N, 72º49’8.3” W |
| 34 | Pomarrosos, Salazar de las Palmas | 13/02/2011 | 1,130 | 7º45’28.9” N, 72º50’33.8” W |
| 35 | Pomarrosos, Salazar de las Palmas | 13/02/2011 | 1,202 | 7º45’33.3” N, 72º50’56.9” W |
| 36 | Pomarrosos, Salazar de las Palmas | 13/02/2011 | 1,320 | 7º45’25.9” N, 72º50’40.2” W |
| 37 | Batagá, Pamplonita | 12/01/2011 | 1,840 | 7º26’12.7” N, 72º38’38.1” W |
| 38 | Batagá, Pamplonita | 12/01/2011 | 1,860 | 7º26’11.3” N, 72º38’39.4” W |
| 39 | Batagá, Pamplonita | 12/01/2011 | 1,891 | 7º26’14.7” N, 72º38’44.1” W |
| 40 | Matajira, Pamplonita | 15/01/2011 | 1,249 | 7º32’45.3” N, 72º37’59.6” W |
| 41 | Matajira, Pamplonita | 15/01/2011 | 1,312 | 7º32’42.3” N, 72º37’53.7” W |
| 42 | Matajira, Pamplonita | 15/01/2011 | 1,385 | 7º32’37.7” N, 72º37’47.7” W |
| 43 | Buenos Aires, Pamplonita | 18/01/2011 | 1,373 | 7º29’9.1” N, 72º37’49.8” W |
| 44 | Buenos Aires, Pamplonita | 18/01/2011 | 1,493 | 7º29’1.7” N, 72º37’42.3” W |
| 45 | Buenos Aires, Pamplonita | 18/01/2011 | 1,568 | 7º29’1.5” N, 72º37’40.3” W |
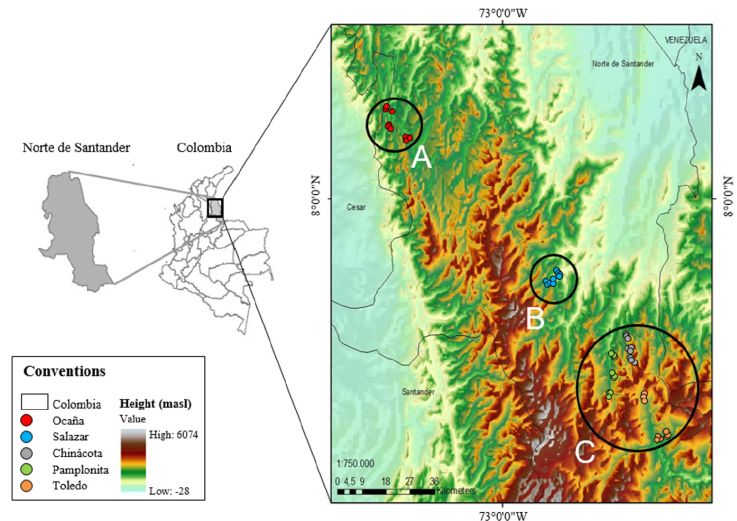
Figure 1 Distribution of the C. moritzianus individuals recorded in Norte de Santander (Colombia, South America). Top of the map, individuals from Ocaña (group A), the middle Salazar region (group B) and in the lower part the municipalities of Chinácota, Pamplonita, and Toledo (group C).
Genomic DNA extraction was performed by macerating the leaves with liquid nitrogen and using the DNeasy plant Qiagen® extraction kit. For the evaluation of the quality and quantity of DNA we used 0.8% agarose gels ran at 90 volts for 30 min in 0.5X TBE (Tris-borate 0045M; EDTA 0.001M). Gels were stained with ethidium bromide (0.5μg/ml). For DNA quantification, extracted DNA was compared with known concentrations of bacteriophage lambda DNA. For amplification by PCR with each RAMs set of primers (CCA, CGA, CT, TG, AG; Muñoz et al., 2008, Table 2), we prepared solutions at a final volume of 12.5 μl containing the following reagents: 1X Taq Buffer, 2.5 mM of MgCl2, 0.2 mM of each dNTP’s, 0.8 mM of each primer, 0.5 U of Taq Polymerase (Promega ™) and 2 μL of extracted DNA. PCR amplification was performed using the following thermal profile: initial denaturation cycle of 5 min at 95 ºC, followed by 37 cycles of 95 ºC for 30 s, hybridization for 45 s at 50 ºC for primer AG, 55 ºC for primers CCA-TG-CT and at 58 ºC for primer CGA, 72 ºC for 2 min and a final extension step at 72 ºC for 7 min.
Table 2 RAMs primers with their corresponding sequences. The following designations for the degenerated sites: H (A or T or C); B (G or T or C); V (G or A or C) and D (G or A or T) (Muñoz et al., 2008). Number of loci and number of polymorphic loci obtained for each primer.
| Primer | Sequence | N° of loci | N° of polymorphic loci |
|---|---|---|---|
| (5’ to 3’) | |||
| CCA | DDB (CCA) 5 | 22 | 20 |
| CGA | DHB (CGA) 5 | 12 | 12 |
| CT | DYD (CT) 7C | 13 | 11 |
| TG | HVH (TG) 7T | 18 | 15 |
| CA | DBDA (CA) 7 | 16 | 15 |
| Total | - | 81 | 73 |
PCR products were observed by electrophoresis in polyacrylamide gels at 7% ran at 150 volts for 1 h 10 min. Staining was performed with ethidium bromide and silver salts as described in standard protocols (Sambrook et al., 1989). Pictures were taken under white and UV light respectively, in a Kodak EDAS 290 transilluminator. A DNA bank composed by the 45 individuals of C. moritzianus was established in the Molecular Biology Laboratory of Universidad Nacional de Colombia, Palmira, which is available at -50 °C (Box 2 / Lot 22). A digital database that includes locations, georeferencing data and concentration of each DNA sample is available.
We recorded and organized the DNA band patterns in a binary matrix: 1 for presence and 0 for absence. A polymorphic locus was considered when the major allelic frequency was inferior to 95%. The genetic similarity between individuals were calculated using the DICE index (Nei, 1978) and carried out a cluster analysis in the SAHN package of NTSYS-pc software v. 2.2 (Rohlf, 2005) using the UPGMA method. To analyze the genetic structure, we performed an analysis of molecular variance (AMOVA) with 999 permutations in the GenAlEx v. 6.5 program (Peakall & Smouse, 2012), based on the allelic frequencies of the 5 populations in 2 levels, within and between the collection areas (2 ways), and the F ST statistic, according to the Wright (1978) scale (0-0.05, low genetic differentiation; 0.05-0.15, moderate; and values greater than 0.15, great differentiation). In order to determine the level of association and proximity between individuals, we performed a multiple correspondence analysis (MCA) over the entire sampled population by using Nei’s genetic distance and the NTSYS-pc software v. 2.2 (Hair et al., 1998; Nei, 1972, 1978; Rohlf, 2005). The expected heterozygosity, percentage of polymorphic loci (95% criterion) and fixation index (F ST, within and between populations) were estimated using TFPGA version 1.3 (Miller, 1997). The genetic distances between geographic groups were estimated using minimum distances and the UPGMA grouping method (Nei, 1972, 1978). Linear geographic distances between the individuals were calculated in DIVA-GIS version 7.5 and correlated with the genetic distances using a partial Mantel test in TFPGA (Hijmans et al., 2012; Miller, 1997; Sokal & Rohlf, 1995). In order to assess whether the altitude is a factor which contributes to population structuring, a hierarchical AMOVA with 999 permutations was performed using GenAlEx v. 6.5, considering the geographical proximity to generate the groups and the altitudinal range of the populations, consequently the variance was partitioned into the following components: among regions (PhiRT), among populations within regions (PhiPR) and within populations in the regions (PhiPT).
Results
DNA concentration ranged between 10 and 30 ng/μl. Extracted DNA had good quality and was suitable for most of the PCR amplification. The 5 RAM primers used in this work, yielded 81 loci with molecular weights between 100 and 1,100 bp. The CCA primer had the highest number of loci, n = 22 and CGA the lowest (n = 12). The CCA primer exhibited the highest number of polymorphic loci (n = 20). The number of polymorphic loci was 73 (Table 2).
The genetic distance dendrogram of the 45 individuals is presented in figure 2. At a coefficient value of 0.66, the individuals are separated into 3 groups: the first group (genotypes = 10) was formed by individuals from Pamplonita, Chinácota and Ocaña. The second group (genotypes = 23) of the dendrogram, shows the largest group of individuals from the municipalities of Ocaña, Chinácota, Toledo, Salazar and Pamplonita and the third group (genotypes = 12) was formed by individuals from Toledo, Salazar, and Pamplonita.
The unbiased genetic distances between individuals ranged between 0.16 and 0.54, values that explain the existence of genetic variability among the individuals of C. moritzianus. The dendrogram grouped the 5 municipalities into 3 distinct groups (Fig. 3). Node 1, corresponding to Ocaña and Chinácota is supported only by 6.17% of the loci, showing a close relationship between the individuals of these 2 localities. Node 2, Salazar and Pamplonita, is supported by 58.02% of the loci; node 3 of Salazar, Pamplonita, and Toledo is supported by 38.27 % and node 4 by 100% of the loci.
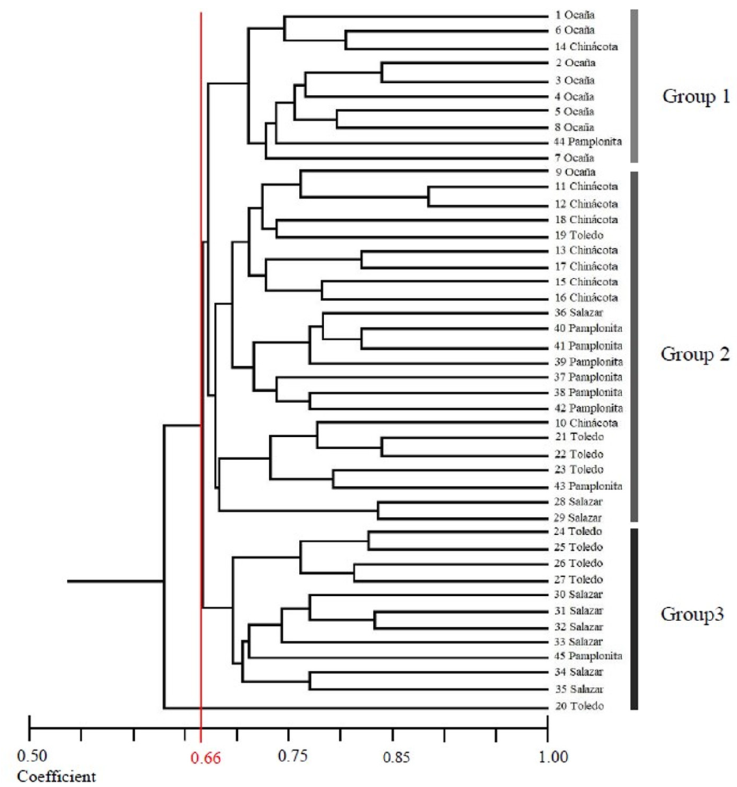
Figure 2 Dendrogram of 45 individuals of Calycolpus moritzianus, based on the coefficient of similarity of Dice (Nei, 1978) and calculated with the combined data of the five RAMs primers.
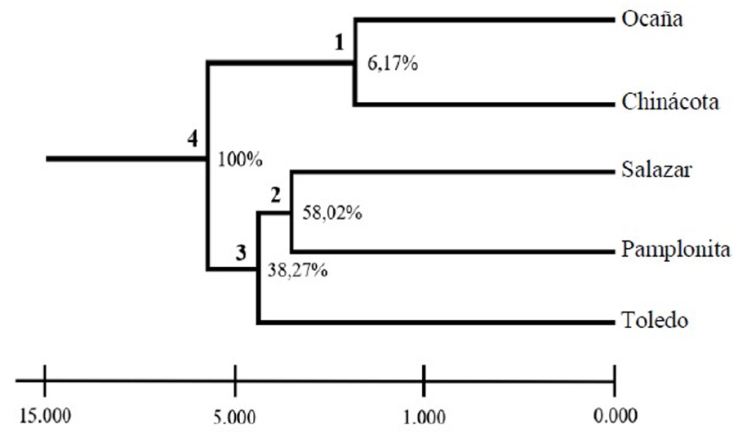
Figure 3 UPGMA from Nei distance (1978) for the 5 populations analyzed and the respective percentages of loci that support each node.
The population of 45 individuals of C. moritzianus in Norte de Santander, showed the highest heterozygosity value for the CGA locus (0.41) and the lowest for the TG locus (0.29); likewise, the highest polymorphic loci percentage was for CGA (100%), followed by CA with 93.80% and TG (77.8%).
When analyzing genetic diversity by individuals, the average diversity among of all individuals of C. moritzianus was found to be 0.35. By municipality, the highest heterozygosity was obtained in Chinácota (0.310), followed by Toledo (0.306), and Pamplonita (0.304), but Ocaña (0.276) and Salazar (0.263) showed a lower diversity. The latter is supported by the molecular analysis of variance (AMOVA), which shows that 14% of the total variation is found between populations, and 86% within populations. Geographical distribution of the individuals indicated 3 groups: group A was composed only by Ocaña, group B only by Salazar, and group C by Chinácota, Pamplonita, and Toledo, with values of F ST = 0.13 (p = 0.01) and SD = 0.024 (Fig. 1). The exact test by population pairs confirms that Ocaña (0.0022, p = 0.01), differs from the other populations.
The geographical groupings indicated above groups A, B and C, was considered in the hierarchical AMOVA. The populations within these groups were nested according to the following altitudinal ranges: < 1,000 m asl, 1,000 - 1,500 m asl, > 1,500 m asl. As a result, a moderate differentiation was obtained between groups or geographic regions (PhiRT = 0.061, p = 0.002), whose contribution to total variance was 6%. No significant differentiation was obtained between populations within groups (PhiPR = 0.027, p = 0.084), for this reason the altitudinal factor contributes to a lesser extent (3%) to population differentiation. Ninety-one percent of the total variation was due to differences between individuals within populations (PhiPT = 0.086, p = 0.001).
The multiple correspondence analysis (MCA) indicated a first group including only individuals from Ocaña (Fig. 4). The remaining populations were included in a second group and were characterized by showing a heterogeneity pattern with 2 distinct sub-clusters: subgroup A with individuals from Pamplonita, Chinácota and Salazar and subgroup B, with individuals from Salazar, Chinácota, Pamplonita and Toledo. The resulting value for the Mantel test was r = - 0.20 (p > 0.05), which indicates no statistical correlation between genetic and geographical distance.
Discussion
This work is a contribution on the population genetic characterization of C. moritzianus in Colombia based on molecular markers. Our combination of primers produced a similar amount of loci as those previously reported for blackberry Rubus spp. (Morillo et al., 2005), the guava Psidium guajava (Sanabria et al., 2006), the gulupa Passiflora edulis f. edulis (Fonseca et al., 2009), the foraging tree Trichanthera gigantea (Posso et al., 2011), the citrus rootstocks (Gallego et al., 2017), the Moringa oleifera tree (Chaves-Bedoya et al., 2017), the oil palm Elaeis guineensis (Castañeda-Cardona et al., 2018), in Zea mays (Alfalahi et al., 2019), and 5 Narcissus species (Mohammed et al., 2020). Given the absence of molecular studies in C. moritzianus, the number of loci generated by the 5 RAMs primers of this work (n = 81) resulted adequate to estimate genetic parameters and other statistical analyzes (Nybom & Bartish, 2000). Despite the fact that RAMs markers are dominant, and therefore do not allow the distinction of homozygous from heterozygous individuals, this multilocus markers, were able to distinguish between individuals and allowed us to differentiate the sampling locations (Hantula et al., 1996; Kremer et al., 2005).
The values of F ST and H e , indicate gene flow and moderate gene diversity between populations, as well as limited population structure, probably due to the short geographic distances between populations of C. moritzianus in Norte de Santander. The distribution of the populations displays a single genetic pool with high variability, settling down 2 groups of individuals, consisting of Ocaña, and the municipalities of Salazar, Chinácota, Pamplonita, and Toledo. The results suggested that the CGA marker is proposed as the most informative for genetic diversity studies in C. moritzianus. The genetic diversity of all populations of C. moritzianus was found to be 0.35 indicating that most of the genetic variability among C. moritzianus locations sampled in this study relies on the individual level rather than on structured populations. This high diversity value could be explained due to the allogamous reproduction of Calycolpus and in general of Myrtaceae, flower structures in this taxonomic group promote gene flow between different organisms (Landrum, 2010; Vasconcelos et al., 2019). This has been also observed in Psidium guajava using RAMs (Sanabria et al., 2006) and RAPD, as guava is a fruit crop with cross pollination and high genetic diversity (Alam et al., 2018). The heterozygosity is higher in Chinácota, Toledo and Pamplonita and lower in Ocaña and Salazar, without necessarily being very low according to Wright (1978) and Balloux and Lugon (2002), and the molecular analysis of variance corroborates this statement, indicating that variation is greater within each one of the municipalities.
The complementary data such as the genetic distances between individuals of C. moritzianus (ranged between 0.16 and 0.54) indicates that, this species, in the sampled region have high genetic diversity. The AMOVA analysis corroborated this, and the results were in accordance with other works in Myrtaceae, in which individuals of P. guajava have high genetic distances of 0.072 and 0.291 in the Valle del Cauca, Colombia (Sanabria et al., 2006).
The values of F ST and the geographic distribution of individuals suggest that Groups B and C can be considered as the same population or genetic pool, but Ocaña conformed one independent group (A), having moderate genetic differentiation (Wright, 1978). According to genetic distance dendrogram analysis (Fig. 2), an individual from Chinácota and another from Pamplonita showed an association with the group made up of individuals from Ocaña, while an individual from Ocaña presented similarity with individuals from Chinácota. This analysis shows a strong tendency for individuals from Ocaña to group together in an independent clade, while individuals from other localities do not present such structure. The MCA also supported the observed tendency for individuals of Ocaña, revealing a separation in 2 populations groups, one located in Ocaña and the second one (with 2 subgroups) that include to Salazar, Chinácota, Pamplonita and Toledo localities (Fig. 4), this is also supported by the AMOVA analysis and F ST comparisons between pairs of populations. However, our data indicated that there is no correlation between genetic and geographical distances, therefore, moderate population structuring is not due to isolation by geographic distance, even if we consider that there is 300 km approximately between the more distant individuals between Ocaña and Toledo.
It has been inferred that one of the main modeling aspects of the associations between the species of Myrtaceae is their tropical origin and the constant adaptation to the different habitats and environmental changes associated with latitude and altitude (Biffin et al., 2010; Rivero, Pacheco et al., 2012). These ecological variables are related with changes in the morphology and flowering patterns in Myrtaceae species, thus generating specialization mechanisms for adaptation to different altitudinal ranges (Biffin et al., 2010; McVaugh, 1968). In the case of C. moritzianus, these ecological variables have been described for the Andean states of Venezuela, where the altitude range for this species is from 600 to 2350 m asl (Pérez, 2007). Very similar values to the ones for the myrtle forests in Norte de Santander, located in a range between 700 to 2,200 m asl (García & Suárez, 2008).
Our altitude measurements for the individuals sampled in this investigation (Table 1) were found within the range known for this species, and there is evidence that some morphological variations have been identified in samples of leaves, flowers and fruits of C. moritzianus in the same 5 municipalities of the present study, observing that there are differences in the morphometric measurements of individuals from Ocaña, compared to Chinácota, Pamplonita, Salazar, and Toledo (Hernández & Medina, 2007; Yáñez et al., 2016). This morphological differences are not enough to support the hypothesis that myrtle populations in Norte de Santander have sufficient morphological variation to propose the presence of varieties of this species (Yáñez et al., 2016). However, it is possible that the interaction species-habitat can be translated into molecular characters that explain the morphological variations within Myrtaceae species (Biffin et al., 2010; Rivero, Salazar et al., 2012).
Our genetic data can shed light on the ideas mentioned previously, which may be feasible because the heterozygosity of C. moritzianus in Ocaña (0.276) is not significantly lower than in the other populations and there is a populational differentiation among Ocaña and the other localities. The hierarchical AMOVA results indicated that there is no differentiation between the populations due to the altitude, which leads us to consider that the genetic and morphological differentiation of Ocaña may not be associated to a process of adaptation of individuals to altitude. However, in the area of this study is the “nudo de Santurbán”, a complex of mountains that occupy 82,664 hectares (ha) between 3,000 and 4,290 m asl in the northeast of the eastern mountain range of Colombia (Morales et al., 2007; Sarmiento et al., 2013), which could be acting as an effective barrier to genetic flow between populations of C. moritzianus. This mountainous area belongs to the Páramos de la Cordillera oriental biogeographic province, established by Londoño et al. (2014), based on angiosperm species composition. The altitudinal distribution of angiosperms in this province and the biogeographic characteristics of the region, indicate that these plants may have geographic isolation between areas of high Andean forest and small paramos, which limits further biotic interchange between areas (Jiménez-Rivillas et al., 2018; Londoño et al., 2014).
The differentiation observed in C. moritzianus from the Ocaña population in the north-eastern Andes of Colombia, may be due to isolation processes by geographic barriers, because orogeny is a factor that determines the geographic ranges in which the species are distributed and influences the potential adaptive of their populations (Eckert et al., 2008). In species with allogamous reproduction such as C. moritzianus (Landrum, 2010), cross-pollination between plants, is expected to show low to moderate population structure, due to increased gene flow, influenced by pollen carryover among flowers and seed dispersal (Bezemer et al., 2016; Harder et al., 2016; Hewitt et al., 2019; Wright et al., 2013). However, geographic barriers, such as the different slopes of the Andes Cordillera range, can promote population differentiation in plants, which has been demonstrated for Rubiaceae (Antonelli et al., 2009; Cueva-Agila et al., 2019), in high Andean Hypericum (Nürk et al., 2013), Espeletiinae (Diazgranados & Barber, 2017), Oreobolus (Gómez-Gutiérrez et al., 2017), Andean Lupinus (Contreras-Ortiz et al., 2018) and Anadenanthera colubrina (Giamminola et al., 2020). Although studies focused on the biogeography of the Myrtaceae family and Calycolpus genera, in the Neotropics are still limited, there is evidence indicating that mountainous environments make up the main areas of diversification in genera such as Myrcia (Santos et al., 2017).
The lack of descriptive ecological information for C. moritzianus and the almost no information on pollinating organisms or dispersers that interact with myrtle forests, do not allow us to assert with certainty this hypothesis. However, an argument supporting this hypothesis is the use of shelters by phyllostomid bats in the Unique Natural Area of the Estoraques, a region close to Ocaña (Suárez & Lizcano, 2011). These results, recorded in Chiroptera feces, seeds of Ficus sp., Eugenia sp., and C. moritzianus, suggesting an association between Artibeus jamaicensis and C. moritzianus, the last one providing a source of food and shelter against predation or adverse weather conditions to the bat (Suárez & Lizcano, 2011). Artibeus are good pollinators and dispersers of Myrtaceae seeds and can generate a significant gene flow exchange for the survival of the myrtle populations (Staggemeier et al., 2010; Tirira, 2017). Although we do not have conclusive evidence of whether this type of dispersal by Chiroptera has an influence on the genetic structure in the myrtle population and the high genetic diversity within populations.
Our results indicated that C. moritzianus is a species with moderate levels of genetic structure and gene flow between populations. Its population structure in the north-eastern Andes of Colombia indicated that this species is a well preserved native genetic resource with a high ecological importance for the country. Finally, considering the commercial potential of the essential oils from C. moritzianus, it is recommended to take advantage (in a sustainable way), of the species and its genetic pool from the 5 evaluated localities and to avoid propagating seeds of a single municipality, preventing the homogenization of the current diversity. Ideally, and together with the local communities, a conservation program must be established that includes germination, sowing and propagation of seeds from the studied populations.











 text new page (beta)
text new page (beta)



