Introduction
The recent coronavirus disease pandemic (COVID-19) is caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), a beta-coronavirus whose RNA genome is ~30kb, while influenza is caused by an RNA virus from the Orthomyxoviridae family, whose genome is ~14Kb. Both viruses infect cells in the respiratory tract, exhibit similar symptoms, and use surface proteins to infect host cells1. Regarding influenza, hemagglutinin (HA) and neuraminidase (NA) are essential for infection. Furthermore, oseltamivir, which is the most widely used treatment against this virus, inhibits the activity of NA by preventing the infection of cells and the release of new viral particles2,3. Conversely, SARS-CoV-2 uses protein S for infection. Two inhibitors directed against this protein were recently developed: E1KC4, which inhibits the fusion of the virus with the target cell, and carmostat mesylate, which blocks the activity of the transmembrane protease serine 2 (TMPRSS2). Both inhibitors result in the lower infectivity of the virus in animal models and human cells in vitro4,5. Both viruses depend on viral RNA polymerase to generate their proteins, which may be another possible therapeutic target1. Nucleoside analogs can insert lethal mutations into the viral code, which lead to their elimination. These analogs have been tested against influenza, but only some are effective due to the virus tolerance to mutations. Recently, the use of the EIDD-2801 nucleoside analog was proposed against SARS-CoV-2 and showed efficacy in human cells in vitro6,7.
SARS-CoV-2 encodes an exonuclease (ExoN) that participates in the genome error correction mechanism, resulting in a reduction in the mutation rate (~10−6 per site per cycle) compared to influenza (~3 × 10−5 per site per cycle), which lacks this mechanism1. This low mutation rate implies that vaccines developed against SARS-CoV-2, as well as the immunity generated in recovered patients, could be long-lasting compared to vaccines and immunity developed against influenza, which becomes obsolete due to strain mutation.
Diseases
Influenza
Influenza is a respiratory disease in humans caused by influenza A (genus influenza virus A) and influenza B (genus influenza virus B) viruses, with an incubation period of 1-2 days, although it can extend to 4 days. Influenza symptoms are characterized by fever, sore throat, runny nose, cough, headache, muscle pain, and fatigue. In severe cases, pneumonia is observed due to the influenza virus or a secondary bacterial infection in the lower respiratory tract8,9. The incidence is higher in pediatric patients and immunosuppressed populations. The most severe presentation is seen in young children, patients over 65 years of age, and individuals of any age with other comorbidities9.
COVID-19
Similar to influenza, COVID-19 is characterized by respiratory symptoms, which include fever, cough, dyspnea, and myalgia in most cases. In severe cases, bilateral lung damage is found. The severity of COVID-19 is associated with advanced age and comorbidities; however, it is not limited to these risk groups10. The Chinese Center for Disease Control and Prevention reported 44,500 confirmed infections: about 80% experienced mild symptoms, while 14% had severe disease with dyspnea and hypoxia; 5% developed critical conditions characterized by respiratory failure, systemic shock, and organ failure. Studies in New York, the epicenter of the pandemic in the United States, reported that of 2634 patients who were hospitalized with COVID-19 between March 1 and April 4, 2020, 14.2% were treated in intensive care units, 12.2% required invasive mechanical ventilation, and 21% died. The mortality of those who required mechanical ventilation was 88.1%11. Although respiratory symptoms dominate the clinical picture of COVID-19, in some patients, viral RNA has been detected in rectal samples even when the nasopharynx test is negative, suggesting possible gastrointestinal infection. Patients with coagulopathies have been reported as well12-14.
Viruses
INFLUENZA VIRUS
These are enveloped RNA (-) viruses with a segmented and relatively small genome of ~14kb, they belong to the Orthomyxoviridae family and are classified as A, B, or C. Influenza A circulates in some mammals, including humans and birds; types B and C also circulate in humans and other species. Influenza A and B are more common than influenza C and cause more severe illness. Influenza C causes a respiratory infection in young children, commonly < 6 years of age, which confers protection when they become adults9. Influenza A is classified by two surface glycoproteins, HA and NA. The specific name of the virus is given based on the proteins it expresses. Therefore, the standard nomenclature is influenza A HxNx, where the x is the number of specific types of HA and NA. Sixteen types of HA (H1-H16) and nine types of NA (N1-N9) have been documented in domestic and wild birds. Three types of viruses (H1N1, H2N2, and H3N2) are responsible for human diseases, and only H1N1 and H3N2 are known to circulate seasonally9.
SARS-CoV-2
This virus was identified as a beta coronavirus a member of the subgenus sarbecovirus and the subfamily orthocoronavirinae. It is a single-stranded RNA (+) virus capable of infecting humans and birds15. There are seven human coronaviruses, of which 229E, NL63, OC43, and HKU1 are associated with the common seasonal cold, while SARS-CoV-1 and MERS-CoV are the causative agents of outbreaks of severe acute respiratory syndrome and the Middle East respiratory syndrome, respectively. The new SARS-CoV-2 is the seventh member of the coronavirus family capable of infecting humans. Genetic analyzes have implicated bats as the natural reservoir for coronaviruses and other animals as potential intermediaries14,16. The SARS-CoV-2 sequence was reported to share 79.6% identity with the previously described SARS-CoV-1 sequence. Furthermore, it has a 96% identity with RaTG13, a bat coronavirus16.
Genome
The influenza A and B viruses genome is ~14kb and contains eight RNA segments encoding 11 proteins. These proteins include three RNA polymerases that function as a complex for the replication of genomic RNA. Since they lack the proofreading function, high error rates of ~10−3-10−4 characterize these RNA polymerases, which leads to the integration of erroneous nucleotides during the replication process17,18. Other translated proteins are HA, which facilitates viral entry, NA, which facilitates the release of new viral particles, nucleoprotein (NP), matrix protein (M1), membrane protein (M2), the non-structural protein (NS1), and the nuclear export protein (NEP)18. HA and NA proteins are the most antigenically variable, as they are located on the surface of the virus and are the main target of neutralizing antibodies8.
The SARS-CoV-2 genome is ~30kb and encodes an RNA-dependent RNA polymerase (RdRp), as well as several structural proteins. Viral RNA is used as a replication and transcription template, and each viral transcript contains a 5 cap structure and a 3 poly (A) tail. On entering the host cell, the RNA genome is transcribed to produce non-structural proteins (nsps) from two open reading frames (ORFs): ORF1a and ORF1b. ORF1a generates the 440-500 kDa polypeptide 1a (pp1a), which is cleaved to 11 nsps. A change in the reading frame, upstream from the ORF1a stop codon, allows the transcription of ORF1b to continue, which generates the 740-810 kDa pp1ab polypeptide cleaved at 15 nsps. This proteolytic cleavage is mediated by the viral proteases nsp3 and nsps that contain a protease domain similar to papain and a type 3C protease domain, respectively19. During replication, anti-sense intermediates (-) are generated, which serve as templates for the synthesis of sense RNA (+) genomic RNA (gRNA) and subgenomic RNAs. Short genomic RNAs (sgRNAs) encode conserved structural proteins such as the spike protein (S), envelope protein (E), membrane protein (M), nucleocapsid protein (N), and various accessory proteins19.
Gene drift and antigenic shift
A mechanism of evolution employed by influenza viruses is the accumulation of mutations in HA and NA, a process known as antigenic drift. In contrast, antigenic shift refers to the exchange of RNA segments between genotypically different influenza viruses, which allows the generation of new antigens, resulting in a new strain or subtype. The three most massive human epidemics in the 20th (1957, 1968) and the 21st (2009) centuries were caused by the antigenic shift between human influenza A virus and influenza A of other species8,17,18. In the case of SARS-CoV-2, it is still soon to have data on this mechanism, but the evidence suggests that this virus originated through a genomic exchange between the human, bat, and pangolin coronaviruses20,21.
Transmission
Human influenza viruses and coronaviruses invade epithelial cells of the upper respiratory tract when viral particles are transmitted through large droplets generated during cough and sneezing by infected individuals. Furthermore, transmission by contact plays an essential role in the spread of the virus. Infected persons, who had touched their mouth and nose or used their hands to cover their sneezes, could eventually be in contact with uninfected individuals who will probably become infected as well by touching their mouth, nose, or eyes9.
Mechanism of Infection
The replication of the influenza virus occurs mainly in the epithelium of the respiratory tract and intestine in birds. The viral life cycle begins when HA binds to sialic acid, which is found on the surface glycoproteins of the target cell. HA from influenza viruses that infect humans preferentially binds to sialic acid joined to oligosaccharides by α-2,6 bonds22. Subsequently, the virus is internalized in an endosome that becomes acidic, allowing HA conformational changes, which induce the fusion of the viral envelope and the endosome membrane. This fusion allows the viral genetic material release in the cytoplasm. The eight viral ribonucleoproteins (vRNPs) are imported into the nucleus of infected cells, where viral RNA transcription and replication occur. Viral RNA transcription produces messenger RNA (mRNA (+)), which is transported to the cytoplasm for translation. The newly translated polymerases (PB1, PB2, and PA) are translocated to the nucleus to increase the rate of viral RNA synthesis, while the viral membrane proteins, HA, NA, and M2, are transported and inserted into the new viral membranes. M1 and NEP bind to vRNPs and leave the nucleus, where recirculating endosomes transport them to the plasma membrane. Once in the plasma membrane, they anchor to the new viral membrane, thus completing the assembly of new virions for their release. Cell death with pathological implications results from constant viral replication8,22.
SARS-CoV-2 uses the same entry receptor as SARS-CoV-1, angiotensin-converting enzyme-2 (ACE-2), but no other coronavirus receptors, such as aminopeptidase N (APN) or dipeptidyl peptidase 4 (DPP4)16. Cell infection by SARS-CoV-2 depends on the previous protein S processing by host proteases. S1/S2 sites, which are processed by furin for their subsequent activation by TMPRSS2, form an exposed loop that contains several arginine residues. The S1 domain binds to the receptor on the target cell, while the S2 domain facilitates the fusion of the viral membrane with the cell membrane. Therefore, the spread of SARS-CoV-2 also depends on the TMPRSS2 protease activity, which could be a therapeutic target23. Recently, the expression of ACE-2 in the lung was described, although it is not limited to this tissue. In 2004, ACE-2 was found on the surface of alveolar epithelial cells and enterocytes of the small intestine. Furthermore, ACE-2 is expressed in endothelial cells of arteries and veins. This receptor distribution could provide information on the possible viral entry routes or other disease manifestations23,24.
Epidemiology
At present, seasonal influenza A (H1N1 and H3N2) circulates in humans. Influenza A of birds or pigs has led to four pandemics since 1918. During pandemics, influenza viruses spread rapidly from the point of origin to the rest of the world in waves of infection25,26. Outbreaks of seasonal influenza occur more frequently during winter in both hemispheres8. Seasonal influenza A and influenza B infections result in ~500,000 deaths annually worldwide27. Pregnant, young, and elderly populations have been considered the most vulnerable populations, with a high risk of infection. The severity and lethality of the disease are attributed to the primary influenza infection, which is characterized by high levels of viral replication in the lower respiratory tract, accompanied by an inflammatory response that leads to a cytokine storm8. In COVID-19, the vast majority of severe cases have been detected in young adults and elderly populations, although infections have also been reported in children. However, it is still too soon to have accurate epidemiological data.
Immune response
Identification of the virus by the innate immune system acts as the first line of antiviral defense. The initial immune response to an influenza virus infection occurs through the secretion of cytokines, induced by the recognition of the viral genomic RNA through various pathogen recognition receptors (PRRs). During the infection of epithelial and endothelial cells and alveolar macrophages, the first wave of cytokines is induced, which includes IFN type I/III, interleukin (IL)-1β, IL-18, tumor necrosis factor (TNF)-α, IL-6, and IL-33. After the release of IFN type I/III, an antiviral response is displayed, with the subsequent production of pro-inflammatory cytokines by dendritic cells, macrophages, neutrophils, and monocytes28. Furthermore, IL-6 participates in neutrophil survival during infection; however, the excessive production of IL-6 and TNF-α correlates with the severity of the disease. In vivo and in vitro, influenza infection also induces the expression of IL-33 by lung epithelial cells, promoting homeostasis, and tissue regeneration through the induction of amphiregulin by regulatory T cells (Treg) and innate lymphoid cells type 2 (ILC2)28.
Coronaviruses are recognized through PRRs: Toll-like receptors (TLR)3, TLR7, TLR9, and RIG-1/MDA5 (retinoic acid-inducible gene-1/melanoma differentiation-associated factor 5). The virus genome recognition generates the activation of the signaling cascades of nuclear factor-kB (NF-kB) and interferon regulatory factor 3 (IRF3), which induce the expression of IFN type I and other pro-inflammatory cytokines29,30. Once secreted, IFN type I activates the JAK/STAT (Janus kinase/Signal transducer and activator of transcription) pathways through the IFN receptor (IFNR). STAT1/2, together with IRF9, induces the expression of stimulated genes by interferon, a response which controls viral infections in early stages29. SARS and MERS have similarities with COVID-19: in mild cases, the immune response is characterized by an interferon antiviral response type I/III and CD4+ Th1 T-cells and CD8+ cytotoxic cell activation, resulting in virus shedding. In severe cases, there appears to be a delay in the antiviral response, with a subsequent increase in the production of inflammatory cytokines due to monocytes and neutrophils infiltration in the lungs, triggering the cytokine storm syndrome. These pro-inflammatory cytokines, such as IL-1, IL-6, IL-12, IL-18, and TNF-α, lead to an increase in vascular permeability and can contribute to respiratory failure29. Another characteristic observed in severe cases is lymphopenia, probably due to direct infection of lymphocytes or to suppression of antiviral responses31.
Adaptive immune response
During infection with influenza, CD4+ T-cells participate in the production of pro-inflammatory cytokines, IFN-γ, and the change of antibodies isotype32, while cytotoxic CD8+ T-cells are responsible for the elimination of cells infected by the virus. Conversely, B-cells are of great importance for the generation of neutralizing antibodies that help to control the infection. The targets of B-cells are mainly HA and NA proteins and, to a lesser extent, NP and matrix proteins33. The antibody response against the influenza virus in humans, without prior exposure, is robust and long-lasting; however, antigenic drift, which mainly affects the HA protein, could cause failures in recognition of the protein by the antibodies34. Since the mucosa of the respiratory tract is the entry site for the virus, secreted antibodies are of great importance in preventing infection, particularly mucosal responses, including immunoglobulin (Ig) A, that are capable of inhibiting virus transmission35. T-cells and ILC2s are activated and secrete a second wave of cytokines that promote virus clearance, homeostasis, and lung tissue repair28.
In recent studies, the presence of protein S-specific CD4+ T cell response was demonstrated in 18 patients with mild, severe, and critical COVID-19. These cells are present in 83% of cases and show an increase in the expression of the activation molecules CD38, human leukocyte antigen complex DR (HLA-DR), and Ki-6736. An interesting finding was the detection of protein S-specific CD4+ T-cells in 34% of healthy individuals seronegative for SARS-CoV-2. However, these CD4+ T-lymphocytes do not present phenotypic markers of activation. However, they are specific for very similar protein S epitopes among human coronaviruses, suggesting a cross-memory in CD4+ T-cells in some individuals (children or young patients who experience a higher incidence of human coronavirus infections)36.
The humoral immune response is essential for virus elimination and the prevention of reinfections. SARS-CoV-2 promotes a potent B-cell response, evidenced by the detection of specific IgM, IgG, and IgA antibodies and neutralizing IgG antibodies. Seroconversion occurs in most COVID-19 patients between 7 and 14 days after the onset of symptoms, and antibody titers persist for weeks after virus clearance31,37. The receptor-binding domain (RBD) of the SARS-CoV-2 protein S is highly immunogenic, and antibodies that bind to this domain could be potent neutralizers, blocking the interaction of the virus with the ACE-2 receptor38. Seroconversion without evidence of reinfections has been detected in COVID-19 patients, suggesting a robust antibody response32. Rhesus monkeys infected with SARS-CoV-2 also resolve the infection and are protected against reinfection39, suggesting protection mediated by B-cells. Studies carried out in common coronaviruses, as well as in SARS-CoV-1 and MERS-CoV, indicate that the specific antibody response decreases over time. In the case of common coronaviruses, it results only in partial protection in case of reinfection. These data suggest that immunity against SARS-CoV-2 might decrease after primary infection; however, studies are required to determine the degree of long-term protection30.
Evasion mechanisms
Although viruses are capable of inducing a potent activation of innate and adaptive responses, sometimes they are capable of evading the immune system, as in the case of influenza viruses, which have several proteins for this purpose. The NS1 protein is capable of binding to RNA, thus preventing the detection and activation of RNA sensors in the cytoplasm, such as RIG-1. Furthermore, NS1 binds to TRIM21 (tripartite motif-containing protein 21) and RNF135 (RING finger protein 135) factors, which are necessary for the activation of RIG-1 after recognition of viral RNA, thus blocking the signaling cascade that produces the expression of interferon and antiviral genes induced by interferon25.
Coronaviruses have evolved more than one mechanism to escape the immune response by avoiding recognition by PRRs. The non-structural proteins (NSP) 10, 13, 14, and 16 of the virus modify viral RNA by methylation and the addition of the guanosine cap at the 5-end. These modifications mimic the mRNA of the host, thus evading the degradation and recognition by PRRs30. SARS-CoV-2 is capable of interfering with the TLRs and IFNRs signaling pathways. The NSP13 protein can interact with the signaling intermediate TBK1 (TANK-binding kinase 1), and NSP15 binds to RNF42/Nrdp1, a TBK1, and IRF3 activator, thus blocking its function40. Other coronaviruses such as SARS-CoV-1 also intervene in downstream signaling pathways of the interferon receptor, decreasing the phosphorylation of the STAT1 transcription factor and preventing its translocation to the nucleus. Whether SARS-CoV-2 follows these mechanisms is unknown, although it appears not to affect STAT1 phosphorylation. The previous studies have shown that SARS-CoV-1 is capable of suppressing IFN release in vivo and in vitro. SARS-CoV-2 is believed to exert a similar effect due to the lack of IFN I/III response, observed not only in primary cells and infected cell lines but also in patients with severe COVID-1941,42.
Furthermore, SARS-CoV-2 NSP9 and NSP10 proteins induce IL-6 and IL-8, which could contribute to the cytokine storm observed in COVID-19 patients43. Elevated levels of IL-6 have been found in patients with COVID-19, showing a correlation with the severity of the disease44. Furthermore, serum levels of IL-6, IL-10, and TNF-α appear to correlate with the lymphopenia observed in patients with severe COVID-19. In contrast, a recuperation in the numbers of T lymphocytes and reduction of pro-inflammatory cytokines was observed in recovered patients45,46.
Complications
Although influenza infection is known as a self-limiting illness, severe manifestations can also be observed. Pneumonia usually occurs with fever, cough, bilateral interstitial infiltrate, hypoxemia, and leukopenia. Deaths from influenza are a consequence of pneumonia that develops during infection, either primary (caused by the virus) or secondary (caused by bacteria), or from exacerbations of cardiopulmonary conditions. However, extrapulmonary complications have been described during influenza infection. The most frequent is secondary pneumonia, and its causative agents are Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, and other Gram-negative bacilli9. Other complications include cardiopathies, Reyes syndrome observed in children treated with aspirin during infection, isolated cases of encephalitis, and acute coronary syndromes. Influenza has also been observed to induce procoagulant activity in cell cultures through the expression of tissue factor9.
In the case of COVID-19, various complications have been observed, which are described below.
Coagulopathies
A recent study examined lungs from patients who died from COVID-19. Seven COVID-19 lungs were compared with lungs from patients who died from acute respiratory distress syndrome (ARDS) secondary to influenza A H1N1 infection47. Lungs of both groups of patients showed alveolar damage and lymphocyte infiltration. However, three specific characteristics were observed in patients with COVID-19: severe endothelial damage associated with the intracellular presence of the SARS-CoV-2 virus, vascular thrombosis with microangiopathy and occlusion of the alveolar capillaries, and the growth of new blood vessels by intussusceptive angiogenesis. Furthermore, the SARS-CoV-2 virus was found within endothelial cells, suggesting that the viral effect, as well as perivascular inflammation, may contribute to endothelial damage47. Other studies have reported the presence of anti-phospholipid antibodies, and elevated levels of Von Willebrand Factor (vWF) and Factor VIII of coagulation in the serum of COVID-19 patients48. Finally, micro-thrombi have also been described in the lungs, lower extremities, hands, brain, heart, liver, and kidney in patients with severe disease11.
SARS-CoV-2 virus receptor, ACE-2, is expressed in endothelial cells, and its primary function is the conversion of angiotensin II (Ang II) into Ang-(1-7). Ang II promotes the formation of reactive oxygen species, while Ang-(1-7) counteracts the effects of Ang II, lowering blood pressure by acting directly on endothelial cells, promoting the synthesis, and release of nitric oxide. ACE-2 and Ang-(1-7) inhibit the formation of arteriosclerotic lesions by protecting the endothelial function and inhibiting the inflammatory response46,49. However, when endothelial damage is present, excessive secretion of vWF is promoted, whose increase in plasma is predictive of prothrombotic complications and organ failure, and is associated with a reduction in survival in the context of severe inflammatory response syndrome50.
Respiratory damage
Respiratory complications seen in patients with COVID-19 are pneumonia and, in severe cases, ARDS. Pneumonia occurs bilaterally: alveoli are filled with fluid, limiting their ability to exchange oxygen and causing respiratory distress, coughing, and other symptoms. If pneumonia progresses, more alveoli fill with fluid from small blood vessels, leading to ARDS. Patients with this syndrome have great breathing difficulties and require mechanical ventilation51,52. Patients with SARS-CoV-2 pneumonia who develop severe respiratory failure (SRF) show hyper-inflammatory responses, with features of immune dysregulation characterized by IL-6 and not by IL-1β. Two key elements are observed in this dysregulation: overproduction of pro-inflammatory cytokines by monocytes and lymphopenia of CD4+ cells, with subsequent lymphopenia of B-cells and depletion of NK cells. Clinical data obtained from patients with COVID-19 show that a cytokine storm and the infiltration of activated immune cells into the lung characterize the early immune response in COVID-19, associated with severe damage to the lung53.
Diagnosis
The diagnosis is based mainly on clinical manifestations; however, samples are taken from the nasopharynx for virus detection. The presence of the virus is evaluated through the reverse transcriptase-polymerase chain reaction (RT-PCR), which is the gold standard for the detection of both viruses. Furthermore, viral cultures are used to characterize possible emerging viruses, as well as rapid antigen detection tests from sputum or nasopharynx samples and serological tests for the detection of IgM or IgG antibodies. However, 7-21 days are required to detect the increase in antibody titers or seroconversion8,9.
Within diagnosed patients, laboratory findings include lymphopenia, elevated levels of inflammatory markers, including C-reactive protein, and increased coagulation cascade factors, including D-dimer. Increased viral load and inflammatory markers positively correlate with the severity of the disease54.
Patient management
Unlike patients infected with influenza, the current management of patients with COVID-19 focuses on controlling infection, supportive care, mechanical ventilation, and managing sequelae and complications54. Suspected COVID-19 patients, who are asymptomatic or have mild disease, are isolated for 14 days from the day of exposure, while patients with severe disease are admitted to hospital for the treatment of hypoxia, respiratory failure, ARDS, and septic shock55. Although observational studies reported that advanced age and comorbidities are risk factors for the severity of COVID-19, severe disease can also occur in young people without pre-existing medical conditions54.
Antiviral treatment
During the flu season, antiviral drugs are used primarily to treat patients with severe symptoms. During a pandemic, when a vaccine is not available, antiviral drugs are essential for treating patients who have been infected and individuals who have been exposed. Two drugs are currently approved for the treatment of influenza56.
M2 ion channel inhibitors
Adamantane derivatives were the first drugs to treat influenza. These derivatives are more effective against influenza A than influenza B. However, these drugs are no longer recommended due to resistance generated by circulating influenza A viruses. The S31N mutation in the RNA segment encoding the M2 protein confers resistance to adamantane derivatives. This event was first seen in influenza A H3N2 virus in 200356.
NA inhibitors
At present, NA inhibitors (NAIs) are the most used for the treatment against influenza. NAIs prevent NA enzymatic activity. There are four NAIs available: oseltamivir (Tamiflu), zanamivir (Relenza), peramivir (Rapivab), and laninamivir (Inavir). Oseltamivir is given orally, zanamivir and laninamivir by inhalation, and peramivir by intravenous route. The administration of these drugs during the first 48 h after the onset of symptoms correlates with a better prognosis. However, in 2007, influenza A H1N1 virus was found to be resistant to oseltamivir. This phenotype is associated with the H275Y mutation in the RNA segment encoding NA. Although these viruses are still sensitive to zanamivir, it becomes necessary to develop new antiviral drugs, such as those found in clinical trials directed against RNA polymerase and HA maturation56.
Viral RNA polymerase inhibitors
In 2014, the viral RNA polymerase inhibitor, favipiravir (T705), was licensed in a restricted manner in Japan. Its use was only allowed for severe cases of influenza with new viruses resistant to the other available therapies. Favipiravir is a competitive inhibitor of viral RNA polymerase and has antiviral activity on various viruses, including influenza. In vivo and in vitro data suggest antiviral activity of favipiravir against influenza A, B, and C, including avian strains. In vitro susceptibility tests demonstrated antiviral activity against viruses resistant to NAIs, with H275Y, R292K, E119V, and D197E mutations. However, studies in animals showed a risk of teratogenicity and embryotoxicity, which explains its restricted use in humans56.
In the case of SARS-CoV-2, no effective drug has been approved, for which developing treatments are being used.
Low molecular weight anticoagulants
Anticoagulant therapy, primarily with low molecular weight heparin, appears to be associated with a better prognosis in severe COVID-19 patients with elevated D-dimer levels. Infection-induced endothelial dysfunction produces excessive thrombin and decreased fibrinolysis, indicating that the patients are in a hypercoagulable state besides COVID-19. In severe COVID-19 cases, hypoxia stimulates thrombosis through an increase in blood viscosity or through hypoxia-inducible transcription factor. As evidence, the development of occlusion and micro-thrombosis in pulmonary small blood vessels in COVID-19 patients has been reported during dissection of the lungs. Therefore, the early application of anticoagulant therapies in severe COVID-19 has been suggested as a possible treatment12.
Monoclonal antibodies
A study in Chinese patients treated with neutralizing anti-IL-6 antibodies reported the recovery of 15/20 patients (75%) in their lymphocyte count and better prognosis. However, a study in Italy informed that only 7 of 33 patients (~21%) improved under this treatment11,53. In another study, the concentration of IL-6 was inversely associated with the expression of HLA-DR in blood. In vitro, tocilizumab treatment in plasma-enriched cell medium restored the expression of HLA-DR in the cell membrane, and the absolute lymphocyte count in six patients53.
Convalescent plasma transfer
Recent studies in patients with COVID-19 confirmed seroconversion through the presence of IgG, IgM, and IgA antibodies, for which passive immunization or transfer of convalescent plasma has emerged as a possible therapy57. It was observed that the time of seroconversion to IgM and IgG was 12 and 14 days, respectively, after the onset of symptoms. Therefore, the collection of plasma three weeks after infection with SARS-CoV-2 was recommended. It was suggested that convalescent plasma therapy led to the improvement of 91/245 COVID-19 patients. Furthermore, in an uncontrolled trial, the transfer of 400 ml of convalescent plasma to five patients with severe COVID-19 caused the resolution of the lung lesions and reduced the severity of the disease and viral load57.
Soluble ACE2
Angiotensin-II converting enzyme (ACE-2) was identified as the entry receptor for SARS-CoV-2. Its expression was detected in the lung, heart, kidney, intestine, and blood vessels, which could explain organ failure observed in infected patients. A recent study demonstrated that the clinical-grade human recombinant soluble ACE-2 (hrsACE2), which has already been tested in Phase I and Phase II clinical trials, can reduce the viral load in vitro in Vero cells. Furthermore, hrsACE2 efficiently inhibits the infection of blood vessels and kidney organoids in the early stages of infection, for which is being promoted as a possible candidate for treating patients with COVID-19 in the early stages of infection58.
Antivirals
A new antiviral method, based on CRISPR-Cas13d, allows the recognition and degradation of the intracellular viral genome, and its resulting messenger RNAs. Furthermore, this method is capable of degrading RNA sequences of highly conserved regions in the genomes of SARS-CoV-2 and influenza A, which inhibits the production of new viruses. Therefore, this tool may be a future antiviral treatment59.
RNA polymerase inhibitors
Recent studies demonstrated that the ribonucleoside analog EIDD-2801 (BD-N4-hydroxycytidine-5-isopropyl ester) possesses broad-spectrum antiviral activity against SARS-CoV-2, SARS-CoV-1, and MERS-CoV. This compound induces lethal mutations in the viral genome, thus preventing its replication. The administration of EIDD-2801 to infected mice with SARS-CoV-2 improved lung function and reduced viral titers, suggesting it as a possible antiviral treatment for COVID-196. Remdesivir also works by inhibiting viral replication. However, EIDD-2801 is 3-10 times more efficient than remdesivir in blocking the replication of SARS-CoV-260.
Major protease inhibitor
The main protease (Mpro or 3CLpro) is the processor of SARS-CoV-2 polyproteins to generate the non-structural proteins (nsp) necessary for virion formation. Two compounds (11a and 11b) were recently designed to target Mpro, and both showed potent inhibitory activity with a subsequent reduction in the infectivity of SARS-CoV-261. An α-ketoamine inhibitor, derived from a previously designed inhibitor modified to increase its half-life in plasma, shows lung tropism when administered by inhalation62. This evidence raises these compounds as possible candidates as therapies against COVID-19.
Fusion inhibitors
The EK1C4 compound can inhibit the fusion of SARS-CoV-2 with target cells. EK1C4 is directed against the HR1 domain of the S2 subunit of protein S. This domain is highly conserved, which reduces the probability of resistance to the compound. Furthermore, the compound proved to be suitable for intranasal administration5. Moreover, protein S protease TMPRSS2 was identified as a therapeutic target due to the requirement for protein S processing to carry out the fusion. Carmostat mesylate efficiently inhibits the TMPRSS2 protease in vitro5, suggesting that both compounds are possible candidates for the treatment of SARS-CoV-2 patients4,5.
Vaccines
The generation of efficient influenza vaccines is challenging due to its mechanisms of drift and antigenic change. However, these vaccines are synthesized and applied on a seasonal basis. Seasonal vaccines are produced by rearranging the six RNA segments of the strain adapted at low temperatures, with the HA and NA RNA segments of the virus specified by the health authorities. However, the objective is to elaborate an effective vaccine against influenza for the long term and against all or most of the antigenic variants8,56.
The development of an efficient and safe vaccine is a priority to end the COVID-19 pandemic. Until the end of May, the World Health Organization published an updated list of candidate vaccines against COVID-19. From this list, ten vaccines are currently in clinical evaluation (Table 1) and 121 in preclinical stages63. On May 22, 2020, a vaccine was announced, which consists of an adenovirus type 5 that expresses glycoprotein S from the Wuhan-Hu-1 strain of SARS-CoV-2. This vaccine was tolerable and immunogenic, obtaining the maximum peak of antibodies after 28 days of vaccination, and the prompt and specific response of T cells by day 14 post-vaccination64. Data suggest that the vaccine is promising. However, Phase III clinical trials are required in larger groups of patients.
Table 1 Candidate vaccines platform in human clinical trials
| Platform | Vaccine type | Developer | Phase |
|---|---|---|---|
| Non-replicative viral vector | ChAdOx1-S | University of Oxford/Astra Zeneca | Phase 2b/3 2020-001228-32 Phase 1/2 2020-001072-15 |
| Non-replicative viral vector | Adenovirus type 5 vector | CanSino Biological Inc/ Beijing Institute of Biotechnology | Phase 2 ChiCTR2000031781 Phase 1 ChiCTR2000030906 |
| RNA | LNP-enveloped mRNA | Moderna/NIAID | Phase 2 NCT04405076 Phase 1 NCT04283461 |
| Inactive | Inactive | Wuhan Institute of Biological Products/Sinopharm | Phase 1/2 ChiCTR2000031809 |
| Inactive | Inactive | Wuhan Institute of Biological Products/Sinopharm | Phase 1/2 ChiCTR2000032459 |
| Inactive | Inactivated by alum | Sinovac | Phase 1/2 NCT04383574 NCT04352608 |
| Protein subunit | Recombinant SARS-CoV-2 glycoprotein nanoparticle vaccine with M matrix as adjuvant | Novavax | Phase 1/2 NCT04368988 |
| RNA | 3-LNP-mRNA | BioNtech/Fosun Pharma/Pfizer | Phase 1/2 2020-001038-36 NCT04368728 |
| Inactive | Inactive | Institute of Medical Biology, Chinese Academy of Medical Sciences | Phase 1 |
| DNA | Plasmid DNA vaccine by electroporation | Inovio Pharmaceutical | Phase 1 NCT04336410 |
DNA, deoxyribonucleic acid; RNA, ribonucleic acid; LNP-mRNA, lipid nanoparticles of messenger RNA.
According to the present information, differences and similarities can be observed between the influenza virus and SARS-CoV-2 (Figure 1). Both viruses infect animals, but for some evolutionary reason or due to the ability to adapt to new hosts, these viruses came to infect humans14. This knowledge is of great relevance, not only to show the actions that can be taken to diminish future infections, especially from unknown viruses, but also to provide insight into how it could alter treatment. The high mutation rate of the influenza A virus (3 × 10−4) gives it advantages to survive within the host. Furthermore, the mutation rate participates in the evasion mechanisms of the immune system, complicating virus elimination, and the durability of vaccines. Finally, the mutation rate can confer the virus resistance to the most commonly used treatment drugs, which represents a significant problem, especially for patients with severe disease1,56.
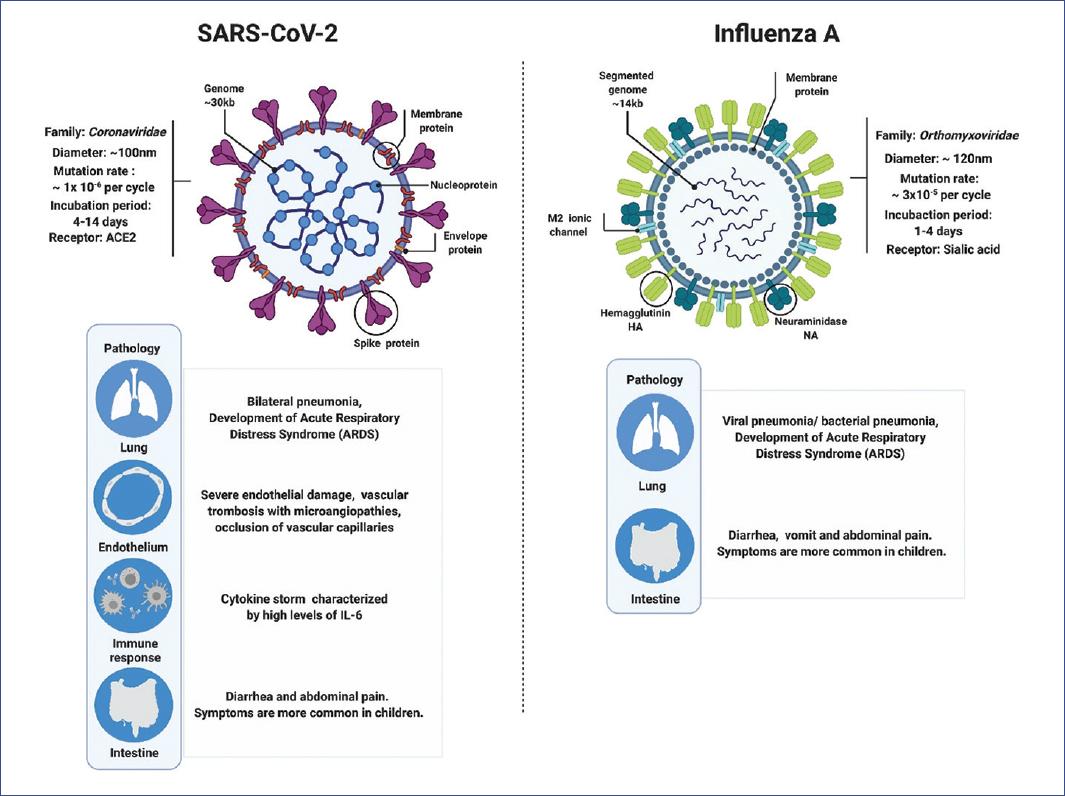
Figure 1 Main differences and similarities between SARS-CoV-2 (severe acute respiratory syndrome coronavirus type 2), which causes coronavirus disease (COVID-19), and influenza A virus, which causes influenza.
In contrast, SARS-CoV-2 has a lower mutation rate (1 x 10-6), which suggests that a vaccine against this virus could be more efficient than those against influenza, as well as the future drugs that will be used as specific treatment against this infection1. At present, multiple compounds are being evaluated for COVID-19 treatment, along with several vaccine candidates (Table 1), although their efficacy is still not clear4,5,12,47,53,5764.
Another critical difference between these two viruses is the response generated by the infection and the observed pathology. Although it is true that initially influenza and COVID-19 appear to be very similar, and the symptoms presented differ very little, the incubation period of SARS-CoV-2 (up to 14 days) is more extended than that of influenza (1-4 days)9,51,52. In both cases, patients develop pneumonia, ARDS, and death in severe cases. However, patients with COVID-19 show severe endothelial damage with thrombosis and microangiopathies in the lungs, which is not observed in patients with influenza47. Other studies support these findings since thrombi and microthrombi have been documented in other parts of the body of COVID-19 patients48. Data suggest that COVID-19 appears to be more vascular than respiratory disease. It is important to note that the receptor used by SARS-CoV-2 is the angiotensin-II converting enzyme (ACE-2). Together with its product (angiotensin 1-7), ACE-2 participates in protecting the endothelium against reactive oxygen species generated by inflammation. However, ACE-2 is unable to perform its protective function by being sequestrated by the virus46. Furthermore, it has been observed that a large number of COVID-19 patients develop the cytokine storm syndrome, dominated by high production of IL-6, TNF-α, IFN-γ, and leukocyte chemoattractants, which generate a wave of amplification of inflammation resulting in systemic damage44.
Both influenza and COVID-19 are dangerous diseases capable of causing death. Based on the knowledge that has been generated, differences and similarities among other viruses can be recognized to allow the identification of the most efficient treatment to fight the current coronavirus pandemic.











 text new page (beta)
text new page (beta)


