Introduction
Bacterial infections cause approximately 17 million deaths per year and are considered a serious global public health concern (de Lima Procópio, da Silva, Martins, de Azevedo & de Araújo, 2012). Despite the discovery of antibiotics and the advances in their production process, the indiscriminate use of these molecules has impacted microbial populations, causing the appearance of strains that are increasingly resistant to multiple drugs and thereby threatening our capacity to treat common infectious diseases (Prestinaci, Pezzotti & Pantosti, 2015). Therefore, the search for new molecules that have broad-spectrum antimicrobial activity against the growing number of multi-resistant strains is crucial (Breijyeh, Jubeh & Karaman et al., 2020).
Actinobacteria are a group of free-living, saprophytic, and filamentous Gram-positive bacteria that are distributed throughout various environments, wherein actinobacteria are chiefly responsible for decomposing organic matter in soil. Among the genera of actinobacteria, Streptomyces is the most studied and considered the most important, since its members produce a large variety of metabolites specialized in functions ranging from the acquisition of nutrients to the inhibition of the growth of competitive organisms (Chater, 2016). Streptomyces members are also known for their ability to produce and release a large variety of secondary metabolites into the environment, many of which possess antimicrobial, antiparasitic, antiviral, and antitumor properties, among others (Qi et al., 2019).
The majority of antibiotics actually in use are originated from Streptomyces bacteria, a group of bacteria of particular importance for medical, agricultural, and veterinary fields (Jones & Elliot, 2017). Approximately 70-80% of all commercially available antibiotics are produced by members of the genus Streptomyces, and they comprise diverse chemical structures and possess diverse biological activities (Wohlleben, Mas, Stegmann & Ziemert, 2016; Al-Ansari, Alkubaisi, Vijayaragavan & Murugan, 2019). The complex life cycle of streptomycetes, their high physiological versatility, and their ability to produce a large variety of secondary metabolites are all reflected in streptomycetes’ genome size. The production of biologically active secondary metabolites is species-specific, and their production depends on the culture conditions and on the nutritional factors in the fermentation medium (de Lima Procópio et al., 2012; Barbuto Ferraiuolo et al., 2021). For instance, antibiotic production by Streptomyces sp. AS4 was upregulated when wheat bran was used as substrate (209 U/g), whereas pomegranate peel supported substantially less antibiotic production (43 U/g). In this sense, the addition of yeast extract (249 U/g), ammonium sulfate (240 U/g), and beef extract (220 U/g) significantly affected antibiotic production (Al Farraj et al., 2020).
Streptomyces is the most important bacterial genus for bioactive compound production, extremely important in biotechnology for the necessity of discovering new compounds to combat emerging resistant microorganisms (Manteca & Yagüe, 2018). The aims of this study were to identify streptomycetes that exert antibacterial activity against plant and human pathogenic bacteria, and to evaluate the antibiotic production of Streptomyces spp. in various culture media.
Materials and Methods
Microorganisms and culture growth
The human pathogens used in this study were Escherichia coli O157:H7, Staphylococcus epidermidis ATCC 1228, Shigella flexneri ATCC 12022, Listeria monocytogenes ATCC 13932, S. aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 33090. The plant pathogen bacteria used were Pseudomonas syringae pv. phaseolicola 1448A, Dickeya dadantii 3937, and Pectobacterium carotovorum. All strains were maintained on Nutrient agar (NA) except Pseudomonas species, which were grown on King B agar (KA). The bacterial inocula were prepared in Mueller-Hinton (MH) broth and King B broth, respectively.
The streptomycete isolates used for this study were obtained from the Conservation of Germplasm Bank of Actinomycetes at the Southeast Unit of CIATEJ. The protocols for culture and storage conditions for the maintenance of Streptomyces are described in the Laboratory Maintenance of Streptomyces Species (Sheperd, Kharel, Bosserman & Rohr, 2010). Repeated streaking onto fresh International Streptomyces Project 2 (ISP2) media was used to reactivate the isolates. To perform agar plug assays and phenotypic characterization, a General Inoculum (GI) of mycelium cells at 0.5 McFarland scale was prepared for each isolate (Evangelista-Martínez, 2014).
Preparation of bacterial pathogenic cultures
Preparation of bacteria inoculums were based on methods of the Clinical and Laboratory Standards Institute (CLSI, 2012). A Petri plate with agar media was inoculated with the pathogens and incubated for 18 h at 37 °C, after which the broth culture medium was inoculated with a loopful of a bacterial colony, mixed, and the bacterial suspension was adjusted to a 0.5 turbidity on the McFarland scale. Then, 0.1 mL of bacterial suspension was dispersed on MH or KA media in a Petri dish. The Petri plates were ready to test for antibacterial activity when the cell suspension was absorbed.
Evaluation of antibacterial activity
An initial screening assay to select antibacterial streptomycetes was carried out with 31 isolates from the Conservation of Germplasm Bank of Actinomycetes. Subsequent assays were performed to evaluate the production of antibacterial compounds under different growth conditions for the selected Streptomyces isolate. Two independent experiments were conducted in quadruplicates.
The agar disk-diffusion method was used to perform a preliminary evaluation to select antibacterial streptomycetes. Initially, 100 µL of the GI suspension was distributed on Petri plates containing ISP2 medium, and the plates were maintained in an incubator for 14 days at 29 °C. Growth of the isolates was then inactivated by placing the plates in a sealed chamber containing chloroform for 30 min. Subsequently, a 9-mm diameter agar disk was removed and deposited on seeded plates inoculated with pathogenic bacteria (as described above), and incubated at 37 °C for 24 h. The diameters of the inhibition halos were measured with a caliper.
An evaluation to observed the effect of culture media on the inhibitory metabolites production was implemented. As mentioned above, 100 µL of the GI suspension were distributed on Petri plates containing the following media: Czapek-Dox agar (CDA), Potato Dextrose agar (PDA), ISP2, ISP2 + 1% (w/v) pancreatic digest casein (Bacto Casitone Difco, ISP2-Cas), ISP9, ISP9 + 1% (w/v) pancreatic digest casein (ISP 9-Cas), NA, Tryptone Yeast Extract agar (TYA), and KA. The inoculated plates were incubated at 29 °C for 14 days. Inhibitory activity was evaluated by the agar-disk diffusion method, as previously indicated.
Inhibitory metabolite production in submerged cultivation and immobilized cell system
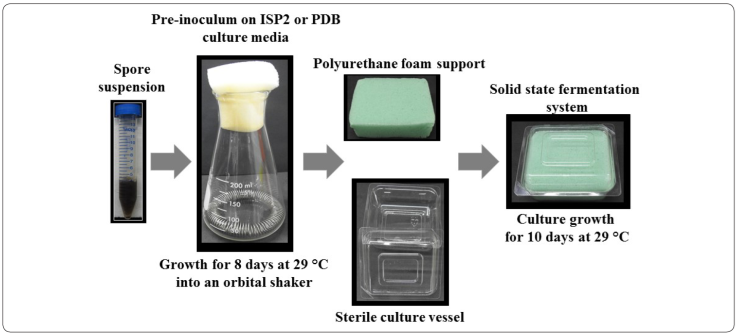
Potato Dextrose broth (PDB) and ISP2 broth media were used. Cultures (50 mL) were grown at 29 °C in 250-mL siliconized flasks with stainless steel springs at 150 rpm in an orbital shaker (Kieser, Bibb, Buttner, Chater & Hopwood, 2000). Cultures were inoculated with a spore suspension that had been washed twice in sterile distilled water and adjusted to an initial optical density of 0.1. Growth was maintained at 29 °C for 8 days.
Solid state cultivation with Polyurethane Foam (PUF) was implemented using PDB and ISP2 broth media. A high density PUF support of L × W × H 12 × 10 × 2.5 cm was washed several times with distilled water and sterilized at 121 °C for 15 min. Then, 50 mL of culture media were inoculated with a spore suspension until the DO450 reached 0.1, and deposited into a sterilized plant cell culture vessel (Phytatray™ II L × W × H 11.4 cm × 8.6 cm × 10.2 cm; Sigma-Aldrich). Thereafter, the PUF was added into each vessel with inoculum under strict aseptic conditions, after which the contents were pressed softly by using a sterile Drigalski spatula in order to allow the PUF to fully and evenly absorb them. The contents were then incubated at 29 °C for 10 days (Figure 1) (Zhu, Smits, Knol & Bol, 1994; Wang & Chen, 2016).
The supernatants (SNs) to be assayed for antibacterial activity were serial-filtered with Whatman® filter paper No. 1, Millipore® filter 0.65 µm and 0.22 µm, and preserved into fresh tubes before storing at −20 °C. Inhibitory activity was evaluated by the disk diffusion method, as previously described. The paper disks used for the diffusion had a 9-mm diameter and were soaked with 30 µL of SN for the assays.
Phenotypic characterization
The Y15 isolate was characterized following the directions given by the International Streptomyces Project (ISP) media on ISP-2 agar (Shirling & Gottlieb, 1966). To evaluate the growth response in the different culture media, 2 μL of GI were inoculated in Petri plates containing the agar media used for antibiotic production. The differentiation steps, which included growth of mycelia substrate and spore production, were observed after a 14-day incubation period at 29 °C. The RAL color chart was used for a visual description of the colonies.
Scanning electron microscopy of isolate Y15
A Petri plate with ISP 2 agar media was streaked with 5 μL of GI and after a 14-day incubation period at 29 °C. An agar block of 14 × 14 mm with an active culture was placed into 2% glutaraldehyde in 0.2 M Phosphate Buffer (PBS, pH 7.2), and left 18 h at 4 °C. Then, the agar pieces were washed in PBS buffer (twice), followed by a 15-min change in 20% ethanol, a 15-min change in 30% ethanol, a 15-min change in 50% ethanol, and a 15-min change in 70% ethanol. The samples were analyzed with an electronic microscope EVO-50 (Carl Zeiss) at the Facultad de Ciencias de la Universidad Autónoma de Querétaro, México.
Molecular identification of the Y15 isolate
The identity of the Y15 isolate was determined based on partial length 16S rRNA gene sequence analysis. The genomic DNA used as a template for Polymerase Chain Reaction (PCR) was prepared from a mycelial cell suspension using the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich®). For 16S gene amplification, the PCR conditions were implemented using the GoTaq® Hot Start Polymerase (Promega) and the oligonucleotides fD1 (5′-CCGAATTCGTCGACAACAGAGTTTGATCC-TGGCTCAG-3′) and rD1 (5′-CCCGGGATCCAAGCTT-AAGGAGG-TGATCCAGCC-3′) (Weisburg, Barns, Pelletier & Lane, 1991). The amplified fragment was directly verified by nucleotide sequence determination of both strands at Macrogen (Seoul, Korea). Sequences were assembled and trimmed using CLC Main Workbench v.6 (QIAGEN ®). The sequences that were analyzed for homology using the BLASTN Suite program and homologous 16S rDNA gene sequencing were retrieved from the 16S rRNA sequences (bacteria and archea) GenBank database (http://blast.ncbi. nlm.nih.gov/). Phylogenetic analysis was carried out at Phylogeny.fr (http://www.phylogeny.fr). The selected sequences were aligned with ClustalW (v.2.1) configured for the highest accuracy. After alignment, ambiguous regions were removed with Gblocks (v.0.91b). The phylogenetic tree was reconstructed using the BioNJ program, and the K2P substitution model was selected for the analysis. The confidence of the grouping was verified by bootstrap analysis (1,000 replications). Partial sequence of 16S rDNA gene sequencing of Streptomyces sp. strain Y15 was deposited in the GenBank database under accession MW714384.
Detection of biosynthetic gene clusters
The biosynthetic genes involved in the production of molecules with antimicrobial properties in Streptomyces sp. Y15 were detected by PCR with specific oligonucleotides. The PCR conditions were implemented using GoTaq® Hot Start Polymerase (Promega). PKS- II gene fragments were amplified with the degenerate oligonucleotides KSα (5′-TSGRCTACRTCAACGGSCACGG-3′) and KSβ (5′-TACSAGTCSWTCGCCTGGTTC-3′) (González, Ayuso -Sacido, Anderson & Genilloud, 2005). The PKS-I fragments were amplified with the degenerate oligonucleotides K1F (5′-TSAAGTCSAACATCCGBCA-3′) and M6R (5′-CGCAGGTTSCSGTACCAGTA-3′). Nonribosomal Peptide- Synthetase (NRPS) gene fragments were amplified with the oligonucleotides A3F (5′-GCSTACSYSATSTACACSTCSGG-3′) and A7R (5′-SASGTCVCCSGTSCGGTAS-3′) (Ayuso-Sacido & Genilloud, 2005). The PCR products were visualized in 1.5% agarose gel prepared in 1X Tris-Borate-EDTA buffer (TBE) and stained with ethidium bromide.
Data analysis
The inhibition halo sizes from the diffusion experiments are expressed as mean ± Standard Deviation (SD). The means for screening of streptomycetes were compared using an analysis of variance (ANOVA) followed by Tukey’s test (p = 0.05). The means for metabolite production by SMF of SSF were analyzed by a two-way ANOVA followed by Tukey’s test (p = 0.05). All statistical analyses were performed with Minitab v.18 (Minitab, LLC).
Results and Discussion
Screening for antibacterial activity of streptomycetes
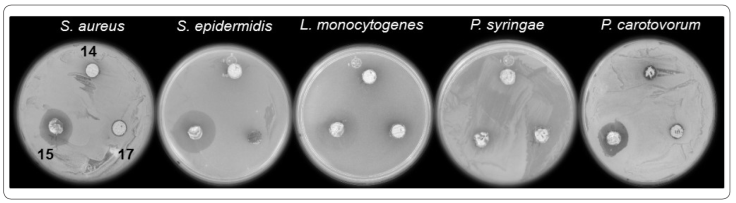
An initial screening to detect antibacterial streptomycetes was essential to select unidentified species. Thirty-one isolates were screened in order to detect inhibitory activity against the bacteria S. aureus, E. coli, L. monocytogenes, S. epidermidis, S. flexneri, P. syringae, and P. carotovorum. Six isolates exhibited clear inhibitory halos surrounding the agar plug-disk from at least two pathogens (Table I). The isolates Y1 and Y41 inhibited four pathogen species (S. aureus, L. monocytogenes, S. epidermidis, and P. carotovorum), while inhibitory halos were detected at all pathogens when agar plug-disk from the isolate Y15 was present (Figure 2).
Table I Antibacterial activity of streptomycetes on bacterial pathogens.
| Isolate | S. aureus | E. coli | L. monocytogenes | S. epidermidis | S. flexneri | P. syringae | P. carotovorum |
|---|---|---|---|---|---|---|---|
| Y1 | 13.27 ± 0.38 | nd | 12.07 ± 0.70 | 18.87 ± 0.74 | nd | nd | 11.80 ± 0.50 |
| Y15 | 18.93 ± 0.67 | 15.20 ± 0.60 | 22.77 ± 1.50 | 26.67 ± 0.83 | 14.17 ± 0.55 | 12.97 ± 0.64 | 20.70 ± 1.21 |
| Y24 | nd** | nd | nd | 14.07 ± 0.72 | nd | nd | 10.80 ± 0.46 |
| Y29 | nd | nd | 20.60 ± 0.78 | 31.87 ± 0.95 | nd | nd | 23.30 ± 1.04 |
| Y40 | 14.40 ± 0.82 | nd | nd | 21.10 ± 0.75 | nd | nd | 24.13 ± 0.68 |
| Y41 | 12.20 ± 0.53 | nd | 13.97 ± 0.42 | 18.70 ± 0.61 | nd | nd | 13.07 ± 0.45 |
| S. lydicus | 12.20 ± 0.80 | nd | nd | 12.53 ± 0.91 | nd | nd | 14.53 ± 1.14 |
| Amikacina* | 30.07 ± 0.35 | 33.60 ± 1.14 | 23.47 ± 1.04 | 36.90 ± 0.70 | 33.60 ± 0.56 | 32.37 ± 0.61 | 41.67 ± 1.35 |
* stock 5 mg/mL, **nd, inhibitory activity non-detected.
Effects of media on antibacterial activity
Secondary metabolites production is known to be greatly influenced by ingredients in the media, especially carbon and nitrogen sources. The effect of culture media on bioactive metabolite production was evaluated on the selected isolate Y15. The isolate was inoculated on Petri plates with nine different culture media, and metabolite production was assessed in S. aureus, E. coli, L. monocytogenes, S. epidermidis, Dickeya dadantii, P. syringae, and P. carotovorum, as previously described. The results showed that the Y15 isolate produced inhibitory metabolites against all of the bacterial pathogens when grown under CDA, PDA, ISP2, ISP9, and ISP9-Cas culture media (Figure 3). Metabolites accumulated in TYA media inhibited six pathogens. When Y15 was grown in ISP2-Cas and KA, five species of bacteria were inhibited, except S. aureus and L. monocytogenes in IPS2 and S. aureus and S. epidermidis in KA. An extreme reduction of bioactive metabolites by Y15 was observed when grown on NA media, wherein inhibitory activity was only exerted against D. dadantii and P. carotovorum. In general, a significantly reduction was observed in the inhibitory halo diameter of the pathogenic bacteria, and fewer species of bacteria were inhibited in culture media containing higher concentrations of a nitrogen source (e.g., peptone from casein, beef extract, and yeast extract as nutrient components of KB, ISP2-Cas, TYA, and NA). Otherwise, culture media with limited or absent nitrogen source supplement showed higher inhibitory halo against bacteria species.
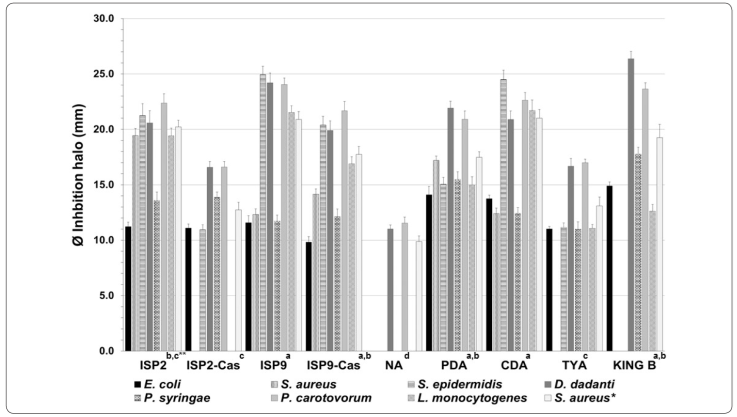
Figure 3 Antibiotic production for Streptomyces sp. Y15 under different culture media growth conditions. Upper images, morphological differentiation of colonies (1-9); 80X total magnification. *S. aureus ATCC 25923. **Means with different letter(s) are significantly different (p < 0.05).
Previous studies acknowledge that carbon and nitrogen sources affect the rate of antibiotic production and morphological differentiation in Streptomyces. A suppression effect of antibiotics production by the carbon source has been reported in recent years. Glucose and other carbohydrates, such as glycerol, maltose, mannose, sucrose, and xylose have been found to interfere with antibiotic synthesis (Sánchez et al., 2010). Similarly, catabolic regulation by nitrogen affected the production of several antibiotics. Downregulation by ammonium has already been observed on macrolides (Tanaka, Taki, Masuma & Omura, 1986) and beta-lactam antibiotics (Braña, Wolfe & Demain, 1985; Castro, Liras, Cortes & Martin, 1985). A high concentration of nitrogen sources (ammonium or amino acids) also suppresses secondary metabolism. For example, production of streptomycin in S. griseus occurs in soybean meal containing in addition L-proline and low concentrations of ammonium salt. Aminoglycoside production was downregulated by ammonium salt, whereas its production was upregulated by nitrate and some amino acids (Shapiro, 1989). Glucose and starch were found to be the best carbon sources for Streptomyces aburaviensis, which produce a broad-spectrum antifungal, whereas NH4NO3 was preferred as a nitrogen source. Supplementing the medium with casein hydrolysate improved both the growth rate and antibiotic titre, but yeast extract resulted in a marked growth rate inhibition (Raytapadar & Paul, 2001).
The manipulation of environment and nutrition for microbial growth has been shown to substantially impact the quantity and diversity of secondary metabolite production, and optimization of this initial step (i.e., microbial culture), may have profound effects on the output and success of a natural products screening program (Shang, Li, Li & Wang, 2012; VanderMolen, Raja, El-Elimat & Oberlies, 2013). Media composition and fermentation conditions (carbon source, phosphate and nitrogen concentrations, pH, temperature), and the supply of specific precursors are the first approaches used to increase the yield in fermentation (Grasso, Martino & Alduina, 2016). Considering the above results, the continued exploration of nitrogen-targeted regulator functions is critical for the eventual implementation of conditions for overproducing or awakening cryptic clusters for secondary metabolite production. Therefore, the composition of the fermentation medium may be an important factor to control antibiotic production by streptomycetes.
Metabolites production by Streptomyces sp. Y15
Submerged fermentation (SMF) is a method of antibiotic production that is affected by diverse problems, including high pollution, low product concentration, and high production cost. An alternative process, which requires a simpler fermentation medium and smaller space, is easier to aerate, has higher productivity, lower waste water output, lower energy requirement, and less bacterial contamination is Solid-State Fermentation (SSF) (Soccol et al., 2017).
Considering that the isolate Y15 grew well and produced antibacterial metabolites in ISP2 and PDB media to a degree comparable to that of carbon- and nitrogen-limited media, an experiment was performed to compare antibiotic production under SMF and SSF (Figure 4). The inhibitory effect of the four SNs on bacterial pathogens were only detected in S. aureus (ATCC 25923), L. monocytogenes, D. dadantii, P. syringae, and P. carotovorum; specifically, inhibitory metabolites that accumulated on SSF exerted the activity in both media. SMF affected the production of antibacterial metabolites, which is related to the incapacity of SN’s to inhibit L. monocytogenes and P. syringae. Statistically significant differences were observed between fermentation conditions (p < 0.05). The results showed differential metabolite production, wherein SSF supernatants exhibiting highest inhibitory activity against the pathogenic bacteria than SMF supernatants.
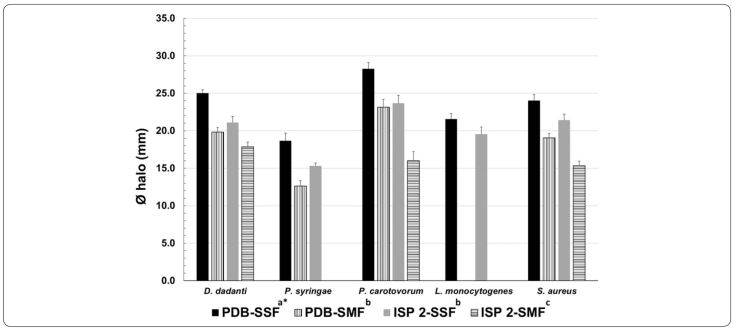
Figure 4 Antibiotic production by Streptomyces sp. Y15 under SMF and SSF. Inhibitory halo of SN evaluated by the paper-disk diffusion assay. *Means with different letter(s) are significantly different (p < 0.05).
It is evident that environmental and physiological signals affect the onset and level of production of each antibiotic. There is a complex interplay of various such signals and regulatory proteins (Kong, Wang, Nie & Niu, 2019). For this, principal factors to consider for efficient antibiotic production are strain selection, medium composition (carbon, nitrogen, and phosphate source availability), growth pH and temperature, oxygen transfer rate, and type of fermentative procedure (Ruiz et al., 2010).
Identification and characterization of Streptomyces sp. Y15
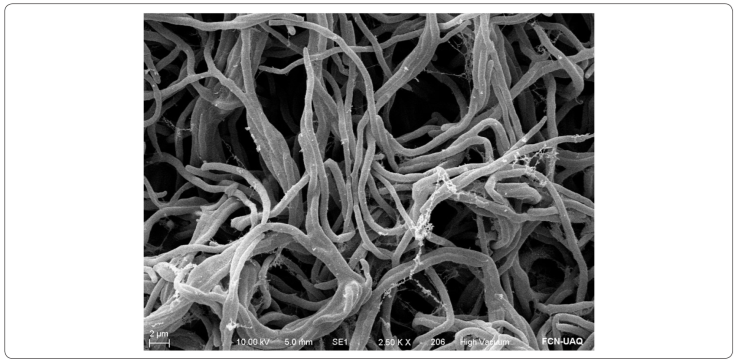
Morphologically, the colonies of the Y15 isolate in ISP2 agar media displayed brown substrate mycelia, white-to-cream aerial mycelia, and spore mass were not evident. This streptomycete has interwoven rope-like hyphae (Figure 5) (Castillo et al., 2005). In addition, some biochemical and physiological features were observed (Table II).
Table II Characterization of Streptomyces sp. Y15.
| Test | Culture media¥ | Growth | Spore | |
|---|---|---|---|---|
| Gram staining | + | ISP 2 | +++ | +++ |
| Casein hydrolysis | + | ISP 2 + casein | +++ | ++ |
| Melanin | - | ISP 9 | ++ | + |
| Growth on ISP2 | Color | ISP 9 + casein | +++ | + |
| Substrate mycelium | Green brown | NA | +++ | + |
| Aerial mycelium | White | PDA | +++ | ++ |
| Spore mass | Beige grey | CDA | ++ | - |
| Pigment production | + | TYA | ++ | + |
| Biosynthetic genes | PCR amplification | King B | +++ | ++ |
| PKS-I | + | |||
| PKS-II | - | |||
| NRPS | - |
¥ The classification of growth and spore production were (+++), excellent; (++), moderate; (+), poor; (-), not detected.
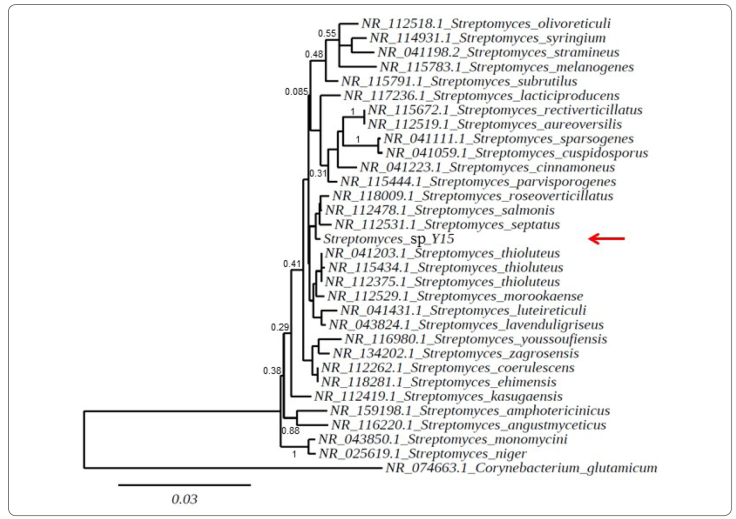
The analysis of the partial 16S rRNA gene sequence (1405 bp) from the Y15 isolate revealed a high similarity (99%) with some Streptomyces sequences retrieved from GenBank. Considering this close relation with other Streptomyces species and the phenotypic features of Y15, the Y15 isolate belongs to the Streptomyces genus. The phylogenetic tree based on the neighbor-joining method showed that the Y15 strain was related to some species capable of producing antimicrobial metabolites (e.g., S. roseoverticillatus, S. salmonis, S. septatus, S. morookaense, and S. thioluteus) (Figure 6). The S. salmonis strain PSRDC-09 produces 14 major volatile organic compounds (VOCs), and l-linalool was the main volatile component, which exerted antagonistic activity on Colletotrichum gloeosporioides revealed as irregular distortions in the fungal hyphae (Boukaew, Cheirsilp, Prasertsan & Yossan, 2021). Streptomyces sp. CMU-MH021, which is 99% identical to S. roseoverticillatus, produced fervenulin, a low molecular weight compound that inhibited egg hatching and increased juvenile mortality of the root-knot nematode Meloidogyne incognita (Ruanpanun, Laatsch, Tangchitsomkid & Lumyong, 2011). S. morookaense AM25 produce a novel cyclic peptide (gloeosporiocide) that exerted activity against the fungal plant pathogen C. gloeosporioides (Vicente Dos Reis et al., 2019). Some streptomycetes in close proximity to Streptomyces sp. Y15 is S. amphotericinicus, which produce amphotericin, a potent antifungal compound, and S. sparsogenes ATCC 25498 produces sparsomicin, an antibiotic with inhibitory activity against Gram-positive and Gram-negative bacteria (Zhang, Zhou, Lou, Wang & Ruan, 2017; Wang et al., 2020).
Detection of biosynthetic gene clusters of secondary metabolites
Genome analyses of actinomycetes revealed that various biosynthetic gene clusters for secondary metabolites are encoded in their genomes and at least about half of the clusters are associated with Non-ribosomal Peptide Synthetase (NRPS) and Polyketide Synthase (PKS) pathways (Nett, Ikeda & Moore, 2009). Polyketides are known to possess remarkable variety in their structure and function (antibacterial, antifungal, anticancer, antiviral, immune-suppressing, and anti-inflammatory activity) (Risdian, Mozef & Wink, 2019). Non-ribosomal peptides are a diverse family of secondary metabolites with a varied property as toxins, siderophores, pigments, antibiotics, immunosuppressants or anticancer agents (Martínez-Núñez & López y López, 2016). These secondary metabolites often show pharmaceutically useful bioactivities, and many have been developed into various drugs (Komaki et al., 2018).
Biosynthetic gene clusters for PKS-I in the genome of Streptomyces sp. Y15 were detected by PCR. The amplified fragment corresponded to ~1400 bp. No DNA fragments of biosynthetic clusters for PKS-II and NRPS were amplified. The conserved modular nature of NRPS and PKS modules allowed detected the gene clusters using designed oligonucleotides on hypervariable regions; the adenylation and ketosynthase domains for NRPS and PKS, respectively. No detection of PKS-II and NRPS in the genome of Streptomyces sp. Y15 could be explained by the fact that these degenerate oligonucleotides may not be beneficial for screening the biosynthetic gene clusters at all streptomycetes isolates. A possible approach would be to used novel oligonucleotides (Amos et al., 2015).
Biosynthetic gene clusters for PKS-I, PKS-II, and NRPS play a fundamental role in the biosynthesis of natural microbial products (Rutledge & Challis, 2015). The members of Streptomyces are widely recognized as antibiotic producers, with genomes containing more than 20 biosynthetic gene clusters for secondary metabolites, representing 5% of their genome (Challis & Hopwood, 2003). In other studies, the presence of at least one of the biosynthetic clusters for PKS-I, PKS-II, or NRPS in actinobacteria was detected by PCR; clusters for PKS-II genes were found to be the most frequent, following by PKS-I and NRPS genes (Lee et al., 2014; Sharma & Thakur, 2020). The absence of PKS-II and NRPS genes does not impair the antibacterial activity of Y15, signifying that different PKS-I clusters or types of bioactive agents may be involved in antibacterial inhibiting activity.
Conclusions
Streptomyces is a bacterium that grows well in nutritionally limited and rich culture media; nevertheless, antibiotic production often requires specific nutrients. For this, optimization of nutritional requirements is fundamental for improving secondary metabolite production. In this genus, secondary metabolism is conditioned by differentiation, which is another aspect that requires special attention given that the onset of morphological differentiation generally coincides with the production of inhibitory metabolites. Therefore, importance should be placed on establishing strategies to activate cryptic biosynthetic gene clusters and discovering novel bioactive compounds.
It was shown that production rate of antimicrobial metabolites was maintained when Streptomyces sp Y15 was grown in culture media containing limited organic nitrogen sources. However, the addition of organic nitrogen compounds in the media decreased the production of antibacterial metabolites.
Detection of biosynthetic gene clusters for PKS-I are generally responsible for producing macrocyclic polyketides (macrolides), characterized by a macrocyclic lactone ring, such as the antibiotics streptomycin, rapamycin, FK506, spiramycin, avermectin, methymycin, narbomycin, and pikromycin. The results obtained in our study indicated a potential for Y15 as a producer of compounds possessing antibacterial properties.
Future work on these antibacterial metabolites should focus on separating and purifying the compounds to probe their activities against drug-resistant bacteria or superbugs. The isolation of these compounds from the crude extracts and chemical analyses would be necessary for structure elucidation.











 text new page (beta)
text new page (beta)