Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista de la Sociedad Química de México
Print version ISSN 0583-7693
Rev. Soc. Quím. Méx vol.46 n.3 Ciudad de México Jul./Sep. 2002
Investigación
Compositional and Conformational Analysis of Sperm Nucleus Basic Proteins from the Bivalve Mollusk Crassostrea virginica
M. Zayil Salazar-Campos, A. Eneas Chavelas-Adame, Enrique García-Hernández1 and Roberto Arreguín-Espinosa*1
Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, México D.F., México 04510. *1Telephone: +52 (55)-5622-4565; Fax: +52 (5) 616 22 17. E-mail: egarciah@servidor.unam.mx; arrespin@servidor.unam.mx
Recibido el 28 de febrero del 2002.
Aceptado el 10 de junio del 2002.
Abstract
We report a study on the basic proteins from sperm nucleus of the bivalve mollusk Crassostrea virginica. This is the first study in its type reported for this specie of wide distribution and high commercial interest. The electrophoretic behavior of total basic proteins was obtained, revealing the characteristic composition of O-group. Using perchloric acid extraction followed by reverse-phase high performance liquid chromatography, a germinal histone H1-2 protein was isolated. An analysis of the circular dichroism spectrum of this protein revealed the lack of regular elements of secondary structure. The H1-2 protein was subjected to a progressive thermal perturbation, observing no abrupt CD-signal change in the scanned temperature range; in contrast, a continuous and significant diminution of optical activity was observed. Overall, these observations indicate that at room temperature the germinal histone is a non-compact intermediate that preserves some secondary structure, and which transits to a more unfolded state upon temperature increase.
Keywords: Crassostrea virginica; histones; sperm nuclear basic protein; composition; purification; circular dichroism.
Resumen
En este trabajo se reporta el estudio de las proteínas básicas nucleares del esperma del molusco bivalvo Crassostrea virginica. Este es el primer estudio en su tipo reportado para esta especie, la cual tiene una amplia distribución geográfica y es de gran interés comercial. De acuerdo al comportamiento electroforético de las proteínas totales obtenidas, se determinó que este organismo posee la composición característica del grupo O. Después de una extracción con ácido perclórico y una separación con HPLC se logró aislar una proteína histona del tipo H1-2. El análisis del espectro de dicroísmo circular de esta proteína revela la ausencia de elementos regulares de estructura secundaria. Posteriormente, esta proteína se sometió a una perturbación térmica sin presentar un cambio abrupto en la señal de dicroísmo circular durante el barrido de temperatura; sin embargo, se observó una disminución de la actividad óptica. De acuerdo a lo anterior, a temperatura ambiente H1-2 posee una estructura intermedia que conserva remanentes de su estructura secundaria, y transita a un estado de mayor desplegamiento conforme la temperatura es incrementada.
Palabras clave: Crassostrea virginica; histonas; proteínas básicas de núcleo de esperma; composición; purificación; dicroísmo circular.
Dedicated to Dr Barbarín Arreguín Lozano
Introduction
In order to be compartmentalized into a cell nucleus of 5-10 µm in diameter, each DNA molecule (∼105 µm long) needs to be folded into a highly compact structure. In eukaryotes, the fundamental unit of the DNA packing system is the nucleo-some. Detailed aspects of the structural organization of the nucleosome have been determined by means of x-ray diffraction analysis [1]. It consists of a protein-DNA complex of ~1:1 mass ratio, with ~150 base pairs wrapped around an octamer of highly basic proteins called histones. About one-fourth of the amino acid residues in histones are arginine and lysine, which aid in shaping an electrostatically-complementary surface towards DNA's negatively charged phosphate groups. The histone octamer is formed of two copies of each histone major class H2A, H2B, H3 and H4. A fifth class of histone, H1, binds internucleosomic DNA, acting as a structural linker that forces nucleosomes to fold into chromatin fiber, and along with other nuclear proteins, into higher-order structures [2]. Based on this wrapping system, the overall compaction of a DNA strand can be 10,000-fold or even greater, yielding ultimately a chromosomic structure.
In early studies on histones, it was thought that they were only passive structural elements that assisted in DNA packing. However, nucleosomes are very dynamic entities, because of the active role of its DNA [3]. Furthermore, histones are preferentially synthesized and suffer specific post-transcriptional modifications depending on the cell type and cycle stage, indicating that DNA activity depends highly on histone regulation [4]. Nowadays, it has become clearly established that nuclear basic proteins play crucial regulatory roles in cellular processes such as gene transcription, replication and repair, and even proteasome specificity [5, 6].
Histones H2A, H2B, H3 and H4 are nearly identical in amino acid composition in all eukaryotes, suggesting that their function has been strictly conserved among species. H4, for instance, is among the most evolutionary conserved proteins known so far, with a maximum interspecies variability below 10 %. In contrast, H1 shows a large degree of variability and structural heterogeneity.
In recent years, information about sperm nuclear basic proteins (SNBP) from organisms of phylogenetically related groups has been gathered [7]. In this regard, seminal studies have been conducted in bivalve mollusks [8, 9]. This information has proved to be valuable not only in providing an appropriate tool to build phylogenetic trees, but also in providing a better understanding of the molecular basis of the function of SNBP.
It is well known that a marked transformation of the SNBP composition takes place during the spermatogenesis process in bivalve mollusks, consisting in a partial or total replacement of somatic histones by protamine-like (PL) proteins [10]. On the basis of the electrophoretic profile exhibited by the germinal set of SNBP in different organisms, Ausió [9] identified five basic PL groups or categories: O, I, II, III and IV. Interestingly, phylogenetically related species tend to gather into the same PL group, indicating that the SNBP structural heterogeneity between taxonomic groups is of a profound evolutionary significance.
We herein report a study of the SNBP from the sperm of the bivalve Crassostrea virginica. As far as we know, this is the first study of SNBP in this widely distributed and highly commercial species. The obtained electrophoretic profile corresponded to the group O type. Using a purification procedure based on perchloric acid extraction and high performance liquid chromatography (HPLC), two different SNBPs were isolated, with a relative abundance of ~30:1. The major component was found to be a germinal H1-2 protein.
Results and discussion
Fractionation and Electrophoretic Analysis of the Total Basic Proteins from C. virginica's Sperm Nucleus.
Figure 1 shows the SDS electrophoretic pattern of total basic proteins extracted from sperm nucleus with hydrocloric acid (lane 1). Lane 2 was loaded with chicken erythrocyte histones (CEH), while standard protein markers were loaded in lane 4. Due to the high abundance of basic residues in SNBP, their specific charge/mass index is larger than that of protein markers, precluding in consequence estimations of the molecular weight of histones through SDS gel electrophoresis.
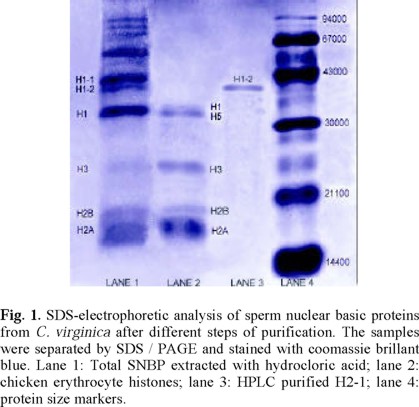
Comparison between lanes 1 and 2 allows identifying the histone types present in C. virginica, as indicated in figure 1. It can be seen that in the gel zone closer to the anode the composition and mobility of SNBP from C. virginica are similar, albeit not identical, to those of CEH. These differences have been shown to be due to variations in the abundance of charged residues and/or molecular weights between somatic and germinal histones [10]. Notably, a number of germinal proteins absent in CEH are clearly identifiable in C. virginica. As indicated in the figure, the germinal H1-like proteins H1-1 and H1-2 are present in the gel, with a lower mobility, evidencing a more basic character of the germinal proteins. Overall, the electrophoretic profile of C. virginica SNBP resembles that of group O, which is characterized by the presence of somatic-like histones (which are absent in the other four PL groups), besides the lower-mobile germinal basic proteins [9]. This group, also known as Pectinidae group, was originally identified in species of the Pectinidae family. Nevertheless, the O-group composition has also been observed in a member of the Ostreidae family, namely C. gigas [9]. As far as we know, C. virginica is the second Ostreidae member showed to belong to the Pectinidae-O PL group.
According to some studies on bivalves, the chromatin organization in somatic and germinal cells is similar, although with variations in the length of the repetitive nucleosome unit. Furthermore, it has been shown that H1-like proteins participate in complexes that either activate or repress specific genes [11]. Thus, it seems likely that the structural differences between somatic and germinal H1-like proteins in group O organisms, and the presence of new specialized basic proteins, determine different strategies of molecular regulation in DNA packing and activity.
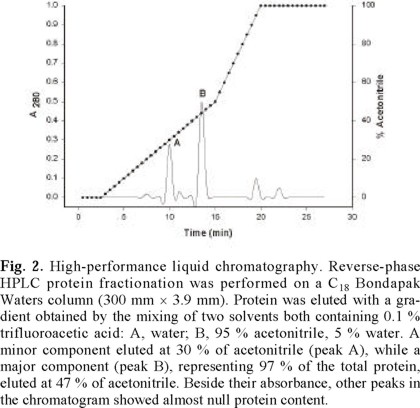
Total SNBP were fractionated with 10 % perchloric acid, and the soluble extract was fractionated by means of reverse-phase HPLC (figure 2). A minor component eluted at 30 % of acetonitrile (peak A), while a major component (peak B), representing ~97 % of the total protein, eluted at 47 % of acetonitrile. Although other peaks appeared occasionally in the chromatographic fractionation of different oyster extracts, their protein content was always marginal in relation to that of the repetitive A and B peaks. In figure 1, lane 3 shows the electrophoretic mobility of the most abundant purified protein component (peak B in figure 2), which clearly corresponds to that of the germinal H1-2 protein in lane 1. In fact, it can be seen that no other band is observable in lane 3, indicating a high degree of purity of H1-2.
Circular Dichroism Measurements. Figure 3 shows the far-UV CD spectrum of H1-2. The spectrum shows a strong negative band centered around 195 nm. The spectrum shape suggests that H1-2 lacks of significant amounts of α-helix elements, which are characteristic of folded somatic H1 [12]. Thus, the CD (circular dichroism) spectrum suggests that germinal H1-2 has little or no structure in solution. The low degree of defined secondary structure in aqueous solution of H1-2 may be due to the electrostatic repulsion between positively charged residues [13]. However, the small positive peak at ~215 nm characteristic of a random coil polypeptide was not observed, suggesting that a small amount of fixed structure could be present in water.
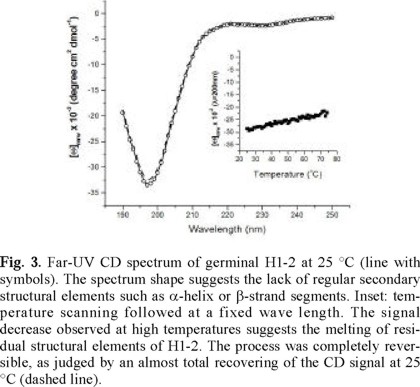
To probe the stability of H1-2, thermal perturbation experiments were performed following the process at a fixed wavelength. As can be seen in the inset of figure 3, the CD signal (at 200 nm) of H1-2 showed a smooth change upon temperature increase. The spectrum at high temperatures was very similar in shape to the one obtained at room temperature, but with an overall intensity decrease of ~25 %. The reversibility of the thermal perturbation was total, as judged by an almost complete recovering of the CD signal when the heated protein solution was re-equilibrated at room temperature (figure 3).
Folded proteins constitute a macroscopic state characterized by a defined and highly-ordered structural ensemble with low dispersion around the average conformation [14]. As a consequence of being very cooperative systems, folded proteins undergo sharp-unfolding transitions in a rather narrow temperature range. In contrast, smooth transitions indicate a non-cooperative behavior, which is characteristic of non-compact conformation states. The thermal perturbation profile depicted by H1-2 is consistent with the picture that it is unfolded at room temperature. Nevertheless, the progressive diminution of the CD signal with temperature increase evidences that H1-2 keeps reminiscent amounts of secondary structure, as it has been observed for other proteins [15]. Overall, this findings indicate that at neutral pH H1-2 is highly unstable, requiring perhaps the presence of its DNA pattern to fold into a compact native structure. Alternatively, the folding of the protein could be achieved by varying solution conditions such as pH or ionic strength. We are currently assaying this issue experimentally to gain insight into the molecular basis of the conformational stability of H1-2.
Experimental
Materials and Methods
Biological material. C. virginica was obtained from commercial suppliers in the market. The sperm collection was carried out with some variations as described before [16]. After opening the shell, the body was rinsed several times with filtered sea water to remove sand and other particles. Single organisms were placed each in a dish with NaCl 0.15M, EDTA 25 mM, pH 8 and 0.1 mM phenylmethylsulfonyl fluoride (buffer A) at 25 °C and gently shaken, until organisms spontaneously released their gametes. The suspension of gametes was collected with the aid of a Pasteur pipette and observed under microscope to check if the material was fast-moving sperm and there was no contamination of other tissues.
Preparation of ripe sperm nuclei. The suspension of sperm in buffer A was filtered through six folds of cheese-cloth and centrifuged at 1500 g for 10 min. The sediment was washed twice with each of the following buffers under the same conditions of centrifugation: NaCl 0.15 M, EDTA 25 mM, pH 8 (buffer A); sucrose 0.25 M, CaCl2 1 mM, MgCl2 5 mM, Tris-HCl 10 mM pH 8, with triton X-100 (buffer B); the latter buffer but without triton X-100 Buffer (C) [17].
Fractionation of Nuclear Basic Proteins. Whole basic proteins were extracted from purified nuclei by stirring overnight at 4 oC with HCl 0.4 N. The crude extract of HCl was suspended in 5 volumes of 10 % perchloric acid, vortexed immediately, and centrifuged at 12000 g for 10 min at 4 oC. The supernatant was precipitated with 6 vol of acetone / HCl at -20 oC. The protein precipitate was recovered by centrifugation at 15000 g for 40 min, washed with cold acetone and dried under vacuum [18].
Quantitation of acid-soluble proteins. Proteins were resuspended in double-distilled water and lyophilized to a volume of 1 mL to concentrate. Protein was quantified using the 280 / 205 nm absorption procedure [19].
High-performance liquid chromatography. Reverse-phase HPLC protein fractionation was performed on a C18 Bondapak Waters column (300 mm × 3.9 mm). Protein was eluted with a gradient obtained by the mixing of two solvents both containing 0.1 % trifluoroacetic acid: A, water; B, 95 % acetonitrile, 5 % water [20]. Acetonitrile from eluted peaks was removed through amicon YM-3.
Circular Dichroism. Far-UV CD spectra were recorded at 25 °C in a JASCO J-720 spectropolarimeter calibrated with (+)-10-camphorsulfonic acid. Measurements were made on a protein solution of ~0.2 mg / mL (0.05 mM phosphates, pH 7.0) in a 0.05-cm cell. Three scanning acquisitions were accumulated and averaged to yield the final spectrum. CD signals are reported as mean residue ellipticity, [Θ]mrw, using a value of 110 for the molecular weight of a mean residue.
Acknowledgments
This work was supported in part by CONACyT (Grant J34303-E) and DGAPA (Grant PAPIIT IN208100 and IN220601).
References
1. Luger, K.; Mader, A. W.; Richmond, R. K.; Sargent, D. F.; Richmond, T. J. Nature 1997, 251, 389. [ Links ]
2. Thoma, F.; Koller, T. Cell 1977, 12, 101-107. [ Links ]
3. Grunstein, M. Annu. Rev. Cell. Biol. 1990, 6, 643-678. [ Links ]
4. Strahal, B. D.; Allis, C. D. Nature 2000, 403, 41-45. [ Links ]
5. Walia, H.; Chen H.-Y.; Sun, J. M.; Holth, L. T.; Davie J. R. J. Biol. Chem. 1998, 273, 14516-14522. [ Links ]
6. Wolffe, A. P. TIBS 1994, 19, 240-244. [ Links ]
7. Ausió, J. J. Biol. Chem. 1999, 274, 31115-31118. [ Links ]
8. Ausió, J.; Van Holde, K. E. Eur. J. Biochem. 1986, 165, 363-371. [ Links ]
9. Ausió, J. Mol. Cell. Biochem. 1992, 115, 163-172. [ Links ]
10. Subirana J. A.; Cozcolluela C.; Palau J.; Unzeta M. Biochim. Biophys. Acta 1973, 317, 364-379. [ Links ]
11. Wolffe, A.P.; Khochbin, P.S.; Dimitrov, S. BioEssays 1997, 19, 247-255. [ Links ]
12. Cerf, C.; Lippens, G.; Ramakrishnan, V.; Muyldermans, S.; Segers, A.; Wyns, L.; Wodak, S. J.; Hallenga, K. Biochemistry 1994, 33, 11079 11086. [ Links ]
13. Vila, R.; Ponte, I.; Jiménez, M.A.; Rico, M.; Suau, P.; Protein Sci. 2000, 9, 627-636. [ Links ]
14. García-Hernández, E.; Hernández-Arana, A.; Zubillaga, R. A.; Rojo-Domínguez, A. Biochem. Mol. Biol. Int. 1998, 45, 761-768. [ Links ]
15. Giancotti V., Russo E., Gasparini M., Serrano D., Del Piero D., Thorne A. W., Cary P. D. and Crane-Robinson C. Eur. J. Biochem. 1983, 136, 509-516. [ Links ]
16. Ausio, J. Comp. Biochem. Physiol. 1986, 2, 439-449. [ Links ]
17. Ausio, J.; Subirana J. A. Exp. Cell Res. 1982, 141, 39-45. [ Links ]
18. Ausio, J. J. Biol. Chem. 1988, 263, 10141-10150. [ Links ]
19. Peterson G. L. Methods Enzymol. 1983, 91, 95-119. [ Links ]
20. Giancotti, V.; Buratti, E.; Santucci, A.; Neri, P.; and Crane-Robinson, C. Biochim. Biophys. Acta 1992, 119, 296-302. [ Links ]





![A Non-sterically Preferred Conformation in Bis-1,4-(2-methyl-4, 5-dihydro-1H-benzo[g]indolyl)benzene](/img/en/next.gif)








