Introduction
Destruction and degradation of native habitat due to anthropogenic land use change are major causes of biodiversity loss worldwide and with increasing negative impacts on developing countries rich in species and ecosystems (Haddad et al., 2015; Torres, Jaeger & Alonso, 2016). Such processes change the spatial configuration and abundance of wildlife populations, which modify dispersal and gene flow patterns at multiple temporal and spatial scales (Didham, 2010). A reduction in dispersal (propagules and individuals) can significantly decrease rates of habitat colonization (Joshi et al., 2006), while a reduction of gene flow between progressively isolated and smaller populations increases the erosion of genetic diversity by the cumulative effects of genetic drift and inbreeding, limiting their adaptive potential (Frankham, 2010). Under climate change, reduced landscape connectivity may hinder the capacity of populations to disperse and establish in suitable areas (Opdam & Wascher, 2004). The synergy between human-induced habitat transformation and climate change is predicted to accelerate species extinctions (Bellard, Bertelsmeier, Leadley, Thuiller & Courchamp, 2012; Opdam & Wascher, 2004). To predict how species may respond and adapt to such changes, it is important to understand how patterns of genetic variation are influenced by the physical and environmental features of the landscape (Pauls, Nowak, Bálint & Pfenninger, 2013). The field of landscape genetics provides a conceptual and methodological framework to advance towards such an understanding, as it examines how gene flow and adaptation occur in heterogeneous landscapes (Manel & Holderegger, 2013).
Mexico is considered a biodiversity hotspot with an exceptional diversity in flowering plants, mammals, reptiles, and amphibians, and it’s among the top five biologically richest countries (Llorente-Bousquets & Ocegueda, 2008; Martínez-Meyer, Sosa-Escalante & Álvarez, 2014). Unfortunately, the country has suffered important rates of habitat fragmentation and degradation in most ecosystems, threatening populations, species, and ecosystem services, which imposes major challenges for its biodiversity conservation under climate change (Moreno-Sánchez, Buxton-Torres, Sinbernagel & Moreno-Sánchez, 2014; Sánchez-Colón, Flores-Martínez, Cruz-Leyva & Velázquez, 2008). There are important advances on the knowledge about genetic diversity of representative taxonomic groups of the Mexican biota, with emphasis on their biogeographic history and population genetics (Piñero et al., 2008). However, less emphasis has been placed on investigating how the landscape context, including anthropogenic landscape change, influence gene flow and adaptation of populations. Such information can considerable inform conservation and management efforts (Keller, Holderegger, van Strien & Bolliger, 2015). To discuss the value of applying a landscape genetics approach for the study of the Mexican biota, the aim of this work is to perform a literature review on current landscape genetics studies in Mexico to highlight major trends of taxonomic groups, habitat types, research questions, and methodological approaches. Towards providing a clear framework, a brief description of this field is introduced, and a qualitative summary of available studies, by grouping them on main research objectives, is presented. Major research gaps have been identified, and six potential areas of research for the Mexican biota are discussed.
Landscape genetics framework
Landscape genetics is an interdisciplinary field that integrates the disciplines of population genetics, landscape ecology, and spatial statistics. The discipline was formally termed in the seminal paper by Manel, Schwartz, Luikart & Taberlet (2003) as the “amalgamation of population genetics and landscape ecology”, which “facilitates our understanding of how geographical and environmental features structure genetic variation at both the population and individual levels”. Landscape genetic studies have extraordinarily increased, in particular, in developed countries and for animal species and temperate ecosystems (Storfer, Murphy, Spear, Holderegger & Waits, 2010). Moreover, advances of molecular and geographic information technologies, combined with new statistical tools and powerful computers, have led to the growth of other subfields such as landscape genomics (investigating tens- to hundreds of loci under selection to identify local adaptation, Schwartz, McKelvey, Cushman & Luikart, 2010), cityscapes genetics (investigating urban environments, Beninde et al., 2016), and seascape genetics (investigating marine ecosystems, Riginos & Liggins, 2013).
The central distinction between landscape genetics and population genetics is the explicit inclusion of spatial and environmental data to test competing hypotheses of determinants of genetic variation, gene flow, and adaptation across heterogeneous landscapes (Holderegger & Wagner, 2006). The temporal and spatial scales are other aspects that distinguish landscape genetics from the field of phylogeography (Manel et al., 2003). Phylogeography focuses on examining the geographic distribution of genealogical lineages to understand geographic historical processes responsible for, e.g., population demographic events, species diversification, refugia zones, and species ranges (Hickerson et al., 2010). In contrast, landscape genetics focuses on more contemporary scales and at finer spatial scales to understand, for instance, the relative influences of landscape configuration and composition (landscape structure) on gene flow (functional connectivity) (Manel & Holderegger, 2013).
Dispersal is a key process for landscape genetics and there are two approaches to infer dispersal through estimates of gene flow. The most common approach is through indirect measures such as population genetic differentiation F ST , which integrates gene flow over several generations (Münzbergová et al., 2013). On the other hand, direct measures, such as parentage analyses, estimate gene flow over one or few mating events and thus are more amenable to detect how gene flow is related to recent habitat changes (Moran & Clark, 2012).
The incorporation of explicitly spatial null models in landscape genetics has become more relevant in recent years, as it is clearer that using a non-spatial null model, such as the island model, is inappropriate (Meirmans, 2012). The most common spatial null model is the inclusion of isolation by geographical distance (IBD), because it assumes that distance per se influences genetic differentiation between individuals or populations, and not the landscape features such as a river acting as a barrier (Holderegger & Wagner, 2008). However, accounting for genetic variation by IBD, while testing for alternative and potential hypotheses, is not a simple task and remains a statistical challenge (Wagner & Fortin, 2016). For instance, the most common statistical approach, the partial Mantel test, which correlates a matrix of pairwise genetic distances (e.g., F ST ) with a matrix of pairwise resistance distances (IBR) (e.g., habitat cover), while controlling for the effect of third matrix of pairwise Euclidean distances (e.g., IBD), has been widely criticized, because it has high type I error rates when variables are not spatially independent (Guillot & Rousset, 2013; Zeller et al., 2016). Although there are alternative statistical analyses (maximum likelihood population effects, spatial generalized linear mixed models, distance-base redundancy analysis), there are no standard methods in landscape genetics, and investigation on the performance of available tests is an ongoing task (Wagner & Fortin, 2016).
Literature Review
A literature search of landscape genetics studies conducted in Mexico between 2003 (the year landscape genetics was officially termed) and March 2017 was implemented using the Web of Science (http://www.isiknowledge.com) with the following key words: landscape genetics and Mexico (n= 50 papers); landscape, genetic diversity, genetic differentiation, gene flow and Mexico (n = 21 papers); landscape, habitat fragmentation, gene flow, genetic diversity, connectivity and Mexico (n = 5). Several papers appeared more than once in these three searches.
Additionally, the Google Scholar search engine was used to ensure a more comprehensive search, which resulted in two papers not retrieved by the Web of Science. Reviews and opinions were excluded. A carefully examination of each of these papers led a total of 20 landscape genetics studies (Table 1).
Table 1 Summary characteristics of 20 landscape genetics studies in Mexico.
| Group | Species | Status | Study Area | Study Objective relating landscape genetics | Marker Type | Reference |
|---|---|---|---|---|---|---|
| Amphibian | Pseudoeurycea leprosa | Threatened | Trans-Mexican Volcanic Belt | Effects of
environmental variables through ENM models on gene flow |
mtDNA sequences, nSSR | Velo-Antón, Parra, Parra-Olea & Zamudio (2013) |
| Amphibian | Ambystoma leorae | Critically endangered | Iztaccihuatl-Popocatepetl National Park | Barriers to gene flow | nSSR | Sunny, Monroy-Vilchis, Reyna-Valencia & Zarco-González (2014) |
| Reptile | Phyllodactylus tuberculosus | Not under concern | Sierra de Alamos Reserve, Sonora | Barriers and effects of vegetation, topography, streams, temperature, and fragmentation on gene flow |
nSSR | Blair, Jiménez Arcos, Mendez & Murphy (2013) |
| Reptile | Urosaurus nigricaudus | Not under concern | Southern Baja California | Barriers to gene flow, and genetic differentiation in continuous vs. fragmented habitat |
nSSR | Munguía-Vega, Jiménez Arcos, Mendez & Murphy (2013) |
| Bird | Pipilo maculatus, Pipilo ocai | Not under concern | Teziutlán and Trans-Mexican Volcanic Belt | Effects of habitat connectivity on introgression at two hybrid zones |
mtDNA sequences, AFLPs | Kingston et al. (2014) |
| Mammal | Phantera onca | Threatened | Sierra Mixe Oaxaca, Sierra del Abra Tanchipa San Luis Potosí | Large-scale and fine-scale genetic structure |
nSSR | Wultsch et al. (2016) |
| Mammal | Liomys pictus | Not under concern | Chamela, Jalisco | Barriers and effects of topography, habitat, precipitation, and streams on gene flow |
nSSR | Garrido-Garduño et al. (2016) |
| Tree | Carica papaya | Not under concern | Los Tuxtlas, Veracruz | Barriers to gene flow, and genetic connectivity of continuous vs. fragmented habitat |
cpSSR and nSSR | Chávez-Pesqueira, Suárez-Montes, Castillo & Núñez-Farfán (2014); Chávez-Pesqueira & Farfán (2016) |
| Tree | Guaiacum sanctum | Endangered | Yucatán Peninsula | Barriers to gene flow | nSSR | Oyama et al. (2016) |
| Tree | Quercus castanea | Not under concern | Lake Cuitzeo, Michoacán | Role
of isolated trees on habitat connectivity by pollen-mediated gene flow |
nSSR | Oyama et al. (2017) |
| Tree | Quercus castanea | Not under concern | Lake Cuitzeo, Michoacán | Effects of habitat fragmentation on seed-mediated gene flow |
cpSSR | Herrera-Arroyo et al. (2013) |
| Tree | Abies religiosa | No under concern | Ajusco Volcano | Ecological and geographic differentiation on local adaptation at outlier loci |
cpSSR, AFLPs | Méndez-González, Jardón-Barbolla & Jaramillo-Correa (2017) |
| Tree | Pinus lumholtzii | Threatened | Durango | Large-scale and fine-scale genetic structure |
AFLPs | Reyes-Murillo et al. (2016) |
| Tree | Picea chihuahuana, Pinus strobiformis, Pseudotsuga menziesii, Populus tremuloides | Endangered (P. chihuahuana) | Durango and Chihuahua | Large-scale and fine-scale genetic structure | AFLPs | Wehenkel, Brazão-Protázio, Carrillo-Parra, Martínez-Guerrero & Crecente-Campo (2015) |
| Succulent | Euphorbia lomelii | Not under concern | Baja California | Constraint of phylogeography on contemporary gene flow |
RFLPs | Dyer, Nason & Garrick (2010) |
| Succulent | Mammillaria pectinifera | Endangered | Tehuacán-Cuicatlán Valley | Barriers to gene flow and population connectivity |
nSSR | Maya-García et al. (2017) |
| Herb | Phaseolus vulgaris | Not under concern | Central and south Mexico | Spatial neutral and adaptive genetic variation at local and continental scales |
SNPs | Rodríguez et al. (2016) |
| Herb | Dieffenbachia seguine | Not under concern | Los Tuxtlas Reserve | Barriers and effects of habitat fragmentation on pollen-mediated gene flow |
Allozymes | Cuartas-Hernández,Núñez-Farfán & Smouse (2010) |
| Epiphyte | Laelia speciosa | Threatened | Lake Cuitzeo, Michoacán | Barriers to gene flow | nSSR | Rojas-Mendez et al. (2017) |
| Parasitic | Psittacanthus schiedeanus complex | Not under concern | Central and south Mexico | Effects of climatic niche and host preferences on genetic differentiation |
cpDNA, nSSR | Ramírez-Barahona, González, González-Rodríguez & Ornelas (2017) |
Source: Author’s own elaboration.
Synthesis of landscape genetics literature
Taxonomic groups, ecosystems, and species conservation status
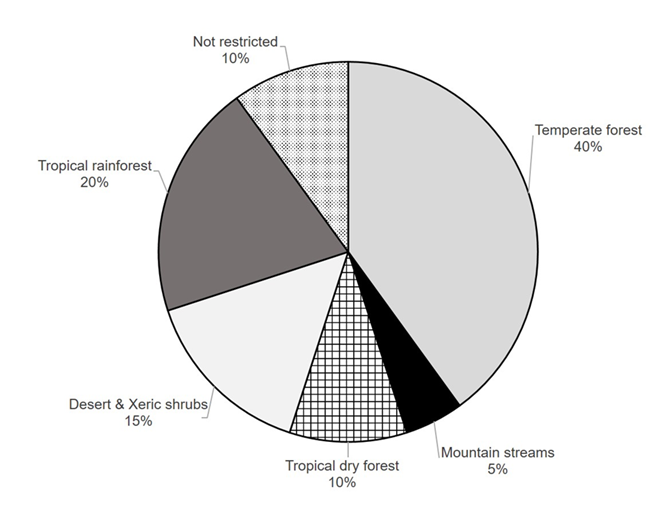
Most landscape genetics studies in Mexico (90%) have been published over the last five years. Of the 20 studies, 65% were conducted in plants, which included 19 species (10 trees, four parasitic plants, two succulents, two herbs, and one epiphyte), whereas 35% were done in animals, which included eight species (two of each amphibians, birds, reptiles, and mammals). A larger number of studies focused on a single species (n = 17), while three studies focused on multiple species (two birds, four coniferous trees, and four parasitic plants). A large proportion of studies focused on temperate ecosystems (40%), such as coniferous forest of pine, oak, and fir species. The second most representative ecosystem was the tropical rainforest (20%), while aquatic ecosystems were scarcely represented (5%) (Figure 1). Sixty seven percent of the species are not within any conservation status, whereas some are under some concern criteria (33%) (Table 1).
Main research objectives
Various studies addressed more than one objective relating to landscape genetics which, for instance, could be estimating linear barriers to gene flow and landscape features, facilitating gene flow. Detection of linear barriers to gene flow was the most popular objective, followed by identification of landscape features facilitating/restricting gene flow. Another proportion of studies focused on evaluating the occurrence of significant patterns of fine-scale genetic structure (FSGS) (Table 1). From these studies, one tested sex-biased dispersal related to habitat fragmentation, while other studies in plants were conducted more as an explorative approach. The next research objective was exclusive to plants, which focused on estimating contemporary estimates of gene flow through parentage analyses for testing differences between fragmented and non-fragmented habitats. Lastly, only two studies in plants were interested to detect potential loci under selection to identify spatial patterns of local adaptation. Major insights of these five objectives will be discussed next.
Landscape barriers to gene flow
All studies found evidence of barriers to gene flow for both animals and plants (Figure 2). However, several studies used barrier detection analysis as an explorative tool, as opposed to hypotheses driven (e.g., Maya-García et al., 2017; Rojas-Méndez et al., 2017). This means that few studies stated a priori hypotheses of specific landscape features, acting as barriers to gene flow, and explicitly acknowledged where to find the hypothesized barriers in the landscape. For instance, Munguía-Vega et al. (2013), for the endemic black-tailed brush lizard (Urosaurus nigricaudus) in a highly fragmented landscape in southern Baja California, hypothesized the occurrence of complete and partial barriers to gene flow on the basis of previous dispersal data. Their results showed that habitat fragments, separated by cleared areas of ~275 m in straight line distance, acted as complete barriers to gene flow, whereas within the scale of dispersal (~175 m), cleared areas and highways separating habitat fragments acted as partial barriers. Two other studies revealed a river as a dispersal barrier: for the leaf-toed gecko (Phyllodactylus tuberculosis) in a tropical dry forest (TDF) in Sierra Alamos in Sonora (Blair et al., 2013), and for the painted spiny pocket mouse (Liomys pictus) in the TDF of Chamela, Jalisco (Garrido-Garduño, Téllez-Valdés, Manel & Vázquez-Domínguez, 2016). For this rodent, a central secondary road traversing the study landscape was not identified as a potential barrier but, in contrast, acted as a dispersal corridor.
Source: Author’s own elaboration.
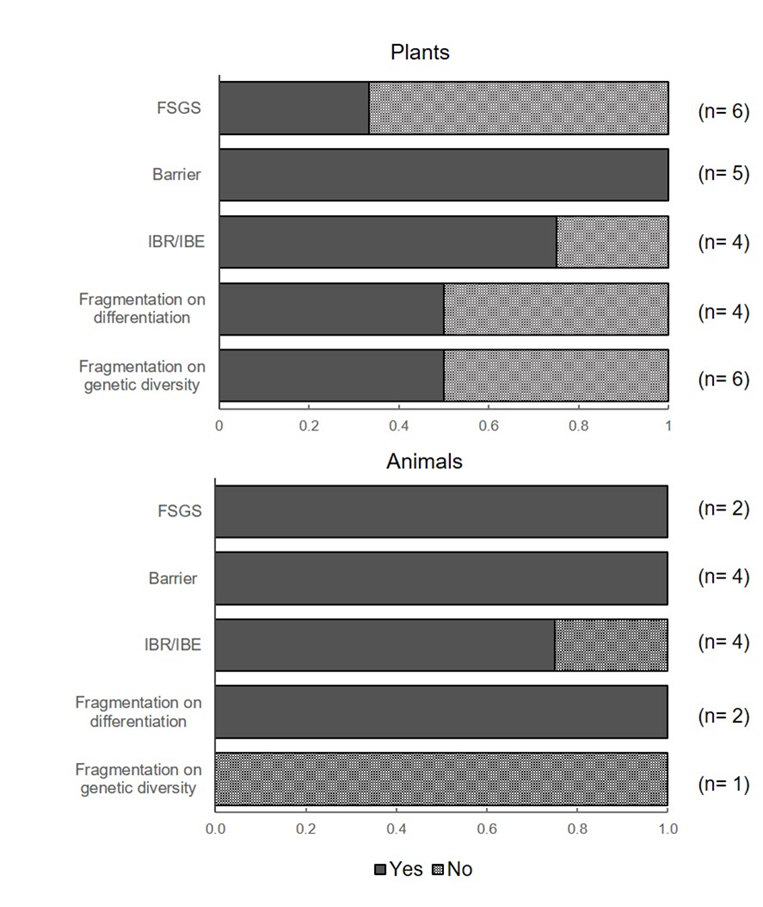
Figure 2 Proportion of species with significant (Yes) and non-significant (No) results for fine-scale genetics studies (FSGS), barriers to gene flow, landscape or environmental features on gene flow (IBR/IBE) and fragmentation effects on genetic diversity and differentiation for plants and animal species.
In plants, detection of barriers to gene flow were more explorative, but all made a link of identified barriers with habitat fragmentation effects. For instance, for an endangered tropical tree (Guaiacum sanctum) across a fragmented landscape in the Yucatán Peninsula, Oyama et al. (2016) detected seven barriers to gene flow, which were associated to habitat fragmentation, although the authors did not discard effects of historical barriers. A similar effect was found for the threatened orchid species Laelia speciosa in the fragmented habitat of the Cuitzeo basin (Rojas-Mendez et al., 2017).
The study by Chávez-Pesqueira & Núñez-Farfán (2016) on the wild cultivar Carica papaya, across its native range, identified historical and contemporary barriers to gene flow using nuclear microsatellites and chloroplast sequences. In addition to barrier detection analyses by boundary detection methods, the authors estimated recent migration rates (first-generation migrants) to evaluate whether detected barriers impeded current gene flow between populations. Their results showed that the Isthmus of Tehuantepec, a putative historical barrier, was not significant, whereas the mountain chains and the fragmented habitat, that had no apparent physiographic features, were detected as barriers to gene flow. The lack of first-generation migrants between populations separated by the mountain chains confirmed them as current barriers to gene flow.
Habitat connectivity, landscape and environmental factors on gene flow
All studies in animals and plants defined a priori hypotheses of potential landscape and environmental features, influencing or limiting gene flow; however, most studies (71%) were not primarily interested in investigating the influence of anthropogenic landscape features. A larger proportion of studies in animals, compared to plants, found that IBR or isolation by environmental (IBE) significantly explained a higher proportion of the genetic variation, as opposed to a model including only geography (IBD) (Figure 2). For instance, Blair et al. (2013), for the leaf-toed gecko, found that IBR models, including forest cover, slope, minimum temperature of the coldest period, and undisturbed habitat, predicted greater gene flow among populations than pure IBD. Similarly, Garrido-Garduño et al. (2016), for the painted spiny pocket mouse, found that precipitation and elevation predicted greater gene flow, whereas vegetation type did not have any significant effect. The authors found significant IBD only for females, which was related to site fidelity and sex-biased dispersal.
Wild relatives of crop species are an important component of food genetic resources, which are threatened under ongoing trends of landscape change. With this motivation, Chávez-Pesqueira et al., (2014) investigated landscape genetic connectivity and genetic diversity of wild populations of Carica papaya. Population connectivity was modeled to account for primary forest, secondary vegetation, agricultural land, and cattle pastures, and with the premise that agriculture and cattle pasture restricted gene flow. Neither IBD nor IBR were significantly correlated with gene flow. However, genetic diversity was significantly low in fragmented populations, which also experienced high genetic differentiation.
Introgression and divergence in hybrid zones has been poorly investigated within a landscape genetics framework, and one of such studies is the work by Kingston et al. (2014) in two towhee species (Papilo ocai and P. maculatus) across two hybrid zones, at Teziutlán and the Trans-Mexican Volcanic Belt (TVB). The authors investigated levels of introgression and divergence by incorporating IBR models of habitat connectivity. The two analyzed hybrid zones showed unique genetic signatures, which were related to the parental populations and to the habitat characteristics of each zone. Interestingly, habitat connectivity, which increased at the intersection between the two hybrid zones, functioned as a dispersal corridor and influenced gene flow dynamics of species hybridization. The insights gained from this study is highly relevant in the light of habitat fragmentation and climate change, since both factors can significantly alter the balance between introgression and differentiation, which would have consequences on species diversity.
Ecological niche modeling (ENM), which combines species occurrence, climatic, land cover, and topographic data to create spatial surfaces of suitable potential habitat, is another approach to model landscape genetic connectivity (Rolland, Lavergne & Manel, 2015). With this framework, Velo-Antón et al. (2013), for the endemic and threatened montane salamander (Pseudoeurycea leprosa), examined how climate and landscape features influence population connectivity across the TVB. However, ENM models did not further explain genetic connectivity after accounting for the effects of IBD. Similarly, investigation of environmental variables through ENM modeling and host preference on population differentiation of mistletoes species was investigated by Ramírez-Barahona et al. (2017), who found that IBE mostly determined genetic structure rather than host specificity.
Disentangling historical and contemporary landscape influences on gene flow is a challenging task in landscape genetics, which is not resolved solely on the basis of marker choice (mtDNA vs microsatellites). This problem is exacerbated in regions of complex biogeographic history such as the Baja California Peninsula, which has undergone a long geological and ecological history, with strong influence on species distribution and genetic structure (Álvarez-Castañeda & Murphy, 2014; Leache, Crews & Hickerson, 2007). To deal with this issue, Dyer et al. (2010) proposed a novel approach based on population genetic networks to remove the effects of phylogeographic history from the contemporary landscape effects on gene flow. They used the stem-succulent Euphorbia lomellii, an arid adapted plant distributed in Baja California, as a study species. Bioclimatic habitat variables were used to model landscape genetic connectivity, while conditional genetic distances (cGD) were derived from population networks. They first fit linear regression models containing the phylogeographic patterns of the species (previously known) as predictor variables expressed as pairwise distance matrices reflecting, for instance, a vicariant event. Residual variation of the best-candidate model was then used as the response variable in linear regressions with bioclimatic distance matrices as predictors. The authors contrasted model inferences between the raw genetic data and the genetic data conditioned by phylogeography. They found, for the conditioned genetic data, that temperature and precipitation significantly predicted gene flow, while elevation was also included as a significant predictor for the raw genetic data. Their results showed that not only variable selection changed between the two data types, but also the relative importance of each predictor.
Fine-scale genetic structure within populations
Spatially restricted dispersal can lead to an increase of relatedness among neighboring individuals, creating a spatial pattern of FSGS. FSGS is often examined through spatial autocorrelation analysis, which test whether the observed genotype of an individual at one site depends on the genotype of a neighboring individual (positive autocorrelation) (Manel et al., 2003). Spatial autocorrelation analyses are useful to determine the spatial scale of a genetic pattern and provide key insights for understanding demographic and ecological processes such as mate choice, sex-biased dispersal, density-dependent effects, and anthropogenic habitat changes (Rico & Wagner, 2016; Vekemans & Hardy, 2004).
Analyzed studies in two species of animals showed significant FSGS, whereas a large proportion of plant species showed no significance (Figure 2). The study by Munguía-Vega et al. (2013), for the black-tailed brush lizard, showed that the spatial scale of FSGS in males within fragmented habitat is markedly larger compared to the spatial scale of FSGS in males for undisturbed habitat. Such findings suggest that habitat fragmentation is importantly changing the dispersal behavior of males, which has direct effects on population dynamics.
FSGS analyses in plants showed varying patterns. Wehenkel et al. (2015) conducted FSGS analyses in four forest tree species in 14 stands in Durango and Chihuahua. For two species, Picea chihuahuana and Pseudotsuga menziesii, significant IBD was found at the broad landscape scale, while none of the four species showed significant FSGS. With a similar approach, Reyes-Murillo et al. (2016), in five stands of Pinus lumholtzii in Durango, found a significant IBD pattern at the landscape scale, but no evidence of FSGS was found, except for two stands. The lack of FSGS in tree species may be related to the high dispersal ability of seeds and pollen, but it may also reflect the influence of ecological variables leading to density-dependent effects. FSGS studies in forest trees are highly relevant to identify the proper spatial scale of seed collection zones to inform reforestation programs.
Contemporary gene flow in anthropogenic landscapes
In plants, landscape-scale genetic connectivity is expected to be sustained by pollen flow as it is likely to travel longer distances and in larger numbers than seeds (Petit et al., 2005). Mexico has suffered important rates of deforestation, and the examination of contemporary pollen-mediated gene flow allows an evaluation of the impact of recent landscape changes. Within this framework, Cuartas-Hernández et al. (2010), for a tropical insect pollinated herb (Dieffenbachia seguine) at Los Tuxtlas Reserve, examined the pollen-pool structure with assignment test in fragmented vs continuous tropical rainforest populations. The authors found that 72% of pollination events occurred with fathers belonging to the same population. For fragmented and continuous populations, progenies were sired by few pollen donors, with moderate pollen immigration into fragments (31%) and into continuous forest (26%). These results suggest that habitat fragmentation has no impact on pollen-mediated gene flow in this species. Using a similar approach, Oyama et al. (2017), for the red oak (Quercus castanea) in a fragmented landscape in the Cuitzeo basin, examined the role of isolated trees in the maintenance of landscape genetic connectivity. Results showed that genetic diversity was similar among adult trees, but the observed heterozygosity was lower in the progenies of isolated trees relative to their mothers. The pollen-pool heterogeneity was similar between trees in forest fragments and isolated trees, and there were no large differences in the effective number of pollen donors. Moreover, ENM analyses revealed an increase of landscape connectivity by the inclusion of isolated trees. This study highlights the importance of isolated trees as stepping stones contributing to genetic connectivity in fragmented landscapes.
Herrera-Arroyo et al. (2013), for the species Q. castanea at the Cuitzeo basin, investigated seed-mediated gene flow by examining adults and seedlings with cpDNA microsatellites. Using a population network approach, which allows the examination of multiple connections among populations simultaneously as opposed to traditional pairwise FST comparisons (Dyer & Nason, 2004), the authors found lower network connectivity in seedlings relative to adults. Network metrics quantifying the number of connections per population and the number of nodes central to network connectivity were significantly low in seedlings. Different from the results of pollen-mediated gene flow for this species (Oyama et al., 2017), this study showed that habitat fragmentation can disrupt seed-mediated gene flow at the landscape.
Gene flow and local adaptation across the landscape
Landscape genomic studies employ hundreds to thousands of loci across the genome, such as Amplified Fragment Length Polymorphism (AFLP) and Single Nucleotide Polymorphism (SNP), to detect those putative loci under selection, or target SNP from specific candidate genes that are putatively involved in adaptation, such as drought stress. Associations between ecological and geographical variables with genetic data allow the examination of adaptive genetic differentiation driven by the environment. The study by Méndez-González et al. (2017) in the temperate forest tree Abies religiosa at the Ajusco Volcano in Central Mexico evaluated landscape patterns of adaptive variation. Twelve putative AFLP loci under selection, which were selected with a combination of statistical approaches to remove false positive loci, showed significant associations with the environment, which explained a much larger proportion of the genetic variation than pure IBD. This study highlights that adaptive divergence in forest trees can occur at the spatial scale of just few kilometers.
Examination of spatial adaptive variation in wild populations of domesticated plants is relevant to improve the genetic resources of crop varieties. Rodriguez et al. (2016), for several wild and domesticated accessions of the common bean (Phaseolus vulgaris) across Mesoamerica and South America, showed that spatial patterns of genetic diversity were high across several localities in Mexico. Putative SNP under selection at candidate functional genes showed strong correlations with climatic variables, including temperature and precipitation. Moreover, varying spatial structure of neutral and adaptive loci among distinct genetic groups revealed the combined effects of limited long-distance dispersal and local adaptation in shaping patterns of genetic structure across the continent.
Research gaps and prospects
Unlike the worldwide trend, in which a large proportion of studies have been conducted in vertebrates (Storfer et al., 2010), in Mexico, landscape genetics studies for plants were more abundant, while methodological approaches were also wider relative to animal studies. However, within these two broad groups, there were obvious gaps in taxonomic groups: for animals, there were no studies in fishes and invertebrates whereas, for plants, most studies were biased towards trees. Ecosystems were also underrepresented. Many studies were for species in temperate ecosystems, mostly due to studies in forest trees. It is striking that aquatic ecosystems were almost absent, with only one study for a freshwater species (Sunny et al., 2014) that tested barriers to gene flow. There was not a single study for marine ecosystems, urban, and semi-urban environments. Moreover, studies explicitly examining genetic diversity and differentiation in fragmented landscapes were few. In the following sections, potential areas of research for the Mexican biota are suggested.
Habitat fragmentation
As pointed earlier, landscape genetics provides new tools to address research questions important for species management in anthropogenic landscapes. Specifically, of the 20 studies reviewed, eight species (six plants and two animals) were examined in the context of habitat fragmentation. Effects of habitat fragmentation on genetic diversity and differentiation were mixed for both animals and plants (Figure 2). Although the number of studies is small to conclude generalizations, the studies in plants showed that genetic diversity was considerably low in fragmented populations for half of the analyzed species. This observation is in agreement with meta-analyses in plants and animals that have revealed the negative consequences of habitat fragmentation on genetic diversity and differentiation (Aguilar, Quesada, Ashworth, Herrerias-Diego & Lobo, 2008; Keyghobadi, 2007). However, more research studies are needed by specifically building a priori competing hypotheses of landscape features on gene flow and genetic diversity. For instance, studies using direct estimates of gene flow with parentage analyses for examining pollen- and seed-mediated gene flow would benefit from incorporating alternative dispersal models accounting for the heterogeneity of the landscape matrix and models reflecting paths of dispersal vectors (Holderegger, Buehler, Gugerli & Manel, 2010). But not only the landscape matrix has an effect on pollen and seed dispersal, other features such as population density, habitat quality, floral diversity, and age structure at the source and destination sites also play an important role (Auffret et al., 2017). The implementation of a gravity population network approach, which allows the integration of both at-site (e.g., population density) and between-site (matrix) characteristics to evaluate the relative contribution of each factor on gene flow would be a potential step forward (DiLeo et al., 2014).
Landscape management
The examination of multiple species and landscapes are other elements to consider in future studies because results from single species and landscapes are unsuitable to translate to other regions and species (Richardson, Brady, Wang & Spear, 2016). Studying multiple co-occurring species give insights of whether common patterns of genetic structuring among species exist in a given landscape, and whether similar environmental features have shaped those genetic patterns. For instance, Fortuna, Albaladejo, Fernández, Aparicio & Bascompte (2009), using population connectivity networks for four co-existing tree species in a fragmented Mediterranean landscape, were able to prioritize functional habitat patches, harboring similar levels of genetic diversity and genetic connectivity in three of the four species. Prioritization of management units to protect the adaptive genetic potential of multiple species is highly desirable (Moritz, 2002).
Moreover, studies in single species preclude the implementation of more comprehensive management strategies, for example, aimed at natural protected areas (NPAs). Mexico has about 11.5% of its territory under some category of NPA. However, the effectiveness of such areas to protect their biodiversity is medium to low, with the exceptions of biosphere reserves (Figueroa & Sánchez-Cordero, 2008). One of the main challenges facing NPAs in Mexico is their low connectivity with other biologically related areas at the landscape and regional scales that would support their long-term functionality, because dispersal and gene flow would not be compromised (Figueroa & Sánchez-Cordero, 2008; Ortiz-Lozano, Gutiérrez-Velázquez & Granados-Barba, 2009). Detection of common landscape features influencing gene flow can be evaluated in comparative landscape genetic studies, which also can identify species-specific responses (Amos et al., 2012; Delaney, Riley & Fisher, 2010; Richardson, 2012). For instance, Amos et al. (2012), for ten woodland-dependent bird species in twelve fragmented landscapes in Australia examined the effects of habitat loss and fragmentation by fitting and ranking competing hypotheses of species dispersal, which allows grouping species responses by their dispersal abilities (mobile vs sedentary). A similar comparative approach can be formulated to identify areas of the landscape that are frequently used across species in order to design functional dispersal corridors between NPAs and their influence zones (Christie & Knowles, 2015).
Another area of improvement in future studies is the quantification of landscape thresholds to gene flow, such as the amount of open areas acting as complete and partial barriers to dispersal, as conducted by Munguía-Vega et al. (2013). For landscape management, quantifying how much of cleared habitats, agricultural land, or forested areas is required to restrict or sustain gene flow would support the delineation of concrete management guidelines (Keller et al., 2015).
Aquatic ecosystems
The study of habitat connectivity in marine environments can be more challenging than in terrestrial ecosystems because of the apparent lack of physical barriers and the high temporal variability of many features; for example, ocean currents (Riginos & Liggins, 2013). Despite the common view that marine species have weak or no genetic structure due to their large population sizes and high dispersal abilities, empirical evidence have shown that marine species can exhibit spatial genetic structure (Selkoe, Scribner & Galindo, 2016). Biogeographic features and environmental variables such as temperature, turbidity, gradients in salinity, ocean currents, upwelling, and resource use in separate habitats during different seasons and ontogenetic stages (larval and adults) are reported to influence spatial genetic structure in marine organisms (Alberto et al., 2010; Amaral et al., 2012; Selkoe et al., 2016).
Mexico has a large diversity of aquatic ecosystems, including inland freshwater environments (e.g., lagoons, rivers, wetlands), coastal (e.g., mangroves, dunes, wetlands, estuaries, coral reefs) and marine habitats (e.g., pelagic and benthonic). The country has 263 coastal municipalities, which experience high urban development because of industrial and tourism growth (Lara-Lara et al., 2008). Anthropogenic activities greatly alter the dynamics of aquatic ecosystems as they can change the chemistry, temperature, turbidity, flow, and nutrients of the water (Selkoe et al., 2016). Climate change is also predicted to strongly impact coastal and marine habitats in Mexico; for example, upwelling may decrease in some zones because of changes in thermal stratification and wind patterns, while in other regions the intensification of winds may promote upwelling (Arroyo, Manzanilla-Naim & Zavalo-Hidalgo, 2011). These changes will impact coastal fisheries as they may present geographic displacements (Arroyo et al., 2011). The application of spatially-explicit models to understand and predict how such processes may affect dispersal and gene flow dynamics in marine environments is, thus, highly relevant.
For freshwater environments, habitat connectivity often respond to the hierarchical organization of these systems, composed of riverine linear networks, varying on gradients of stream flow, volume, and nutrient composition, but it can also be fragmented by features such as dams, cascades, and mountains (Hopken, Douglas & Douglas, 2013; Lean, Hammer, Unmack, Adams & Beheregaray, 2017). Semi-aquatic organisms, such as insects and turtles, are likely to move between permanent water bodies, ephemeral ponds, and terrestrial linear features that differ in dispersal permeability (Murphy, Dezzani, Pilliod & Storfer, 2010). Moreover, temporal and spatial oscillations in habitat permeability, due to flood or drought events, can largely influence demographic and gene flow metapopulation dynamics of plant and animal species (Watts et al., 2015). Landscape genetics of aquatic and semi-aquatic organisms is feasible by building spatially-explicit habitat connectivity models, incorporating the spatial hierarchical organization of aquatic systems, the temporal and spatial variability of environmental features, and physical barriers to dispersal (Selkoe et al., 2016).
Urban environments
Urbanization is considered an important driver of landscape change, which often reduces the diversity and abundance of native biota, while promoting (intentionally by humans or not) the spread and establishment of non-native species (McKinney, 2006). Empirical evidence reveals that urbanization can significantly reduce gene flow and genetic diversity among populations within cities (Lourenço, Álvarez, Wang & Velo-Antón, 2017; Munshi-South, Zolnik & Harris, 2016). On the other hand, green areas, such as public parks, gardens, and residential yards harboring native and non-native flora, can provide an important ecological value, acting as refuges and food reservoirs for pollinators or as stepping stone habitat for species dispersal (Su et al., 2015).
Urban and peri-urban ecological studies across several cities in Mexico have shown that green areas within the city can harbor a considerable number of native and non-native species (e.g., Mexico City: MacGregor-Fors, 2008; Oaxaca: Pablo-López & Díaz-Porras 2011; Querétaro: Pineda-Lopez, Febvre & Martínez, 2010). Differences on species diversity among urban areas has been associated to the degree of contiguous urbanization (MacGregor-Fors et al., 2015). A next research step to conduct in those cities that have already species inventories would be to investigate spatial patterns of genetic connectivity among wildlife populations inhabiting green areas. Urban landscape genetics of key native species can assist a sustainable urban development planning, since the ideal goal would be to integrate green urban cities as another component of the biodiversity conservation across the landscape (McKinney, 2006).
Species interactions
Landscape community genomics aims at understanding how neutral and adaptive genetic variation of interrelating species is determined by biotic and abiotic factors across the landscape (Hand, Lowe, Kovach, Muhlfeld & Luikart, 2015). This perspective stresses that species interactions such as competition, mutualism, disease spread, and hybridization are context-dependent and with direct influences on genomic variation (Hand et al., 2015; James, Coltman, Murray, Hamelin & Sperling, 2011). The study by Kingston et al. (2014) on species hybridization is a good example of the type of research interest within landscape community genomics. In Mexico, there are many taxonomic groups (e.g., oaks, pines, butterworts, hummingbirds, lizards) in which hybridization has played a significant role on species diversification. Thus, studying hybrid zones by incorporating the effects of the spatial context would advance our understanding on the maintenance of reproductive barriers in the occurrence of gene flow. Moreover, investigating range shifts of hybrid zones under climate change would provide insights for understanding how biotic and abiotic factors influence negative (maladaptation) or positive (adaptive variation) outcomes of introgression in interacting species (Taylor, Larson & Harrison, 2015).
Other relevant processes to address with this framework are disease spread and species invasions, such as the rapid spread of the dwarf mistletoe in central Mexico (Queijeiro-Bolaños, Cano-Santana & Castellanos-Vargas 2013), pine beetle outbreaks in mountain regions (e.g., TVB, Sierra Madre Oriental and Sierra Madre del Sur [Salinas-Moreno, Ager, Vargas, Hayes & Zu, 2008]), and spread of mosquito-borne diseases (e.g., malaria, Zika and Chikungunya virus) across Mexico. Disease spread, and species invasions have negatively impacted native biota and public health, which can significantly increase under climate change. Host-pathogens interactions share strong spatial dependencies with landscape features that can influence rates of dispersal and contagion (McCallum, 2008). In urban environments, for instance, spatially-explicit hypotheses have been tested to predict the spread of rabies virus carried by raccoons in cities across the United States (US) and Canada (Côté, Garant, Robert, Mainguy & Pelletier, 2012; Cullingham, Kyle, Pond, Rees & White, 2008). Another relevant work is the study by Medley, Jenkins & Hoffman (2015), who investigated the spread of the Asian tiger mosquito (Aedes albopictus), responsible for dengue, West Nile and Chikungunya virus across the US. This work explicitly investigated the contribution of dispersal by human transportation and natural features of the landscape. Their results revealed that long distance dispersal via highways facilitates gene flow, while at smaller spatial scales, forest and agricultural land restricted gene flow. The application of this framework in combination with computer simulations would greatly inform the design of control measures for zoonotic diseases.
Adaptation to climate change
Mexican ecosystems such as cloud forest, coniferous forest, semi-deciduous forest, and alpine ecosystems are highly threatened by climate warming because most of their species are adapted to cold and wet conditions (Gómez-Mendoza & Arriaga, 2007; Ramírez-Amezcua, Steinmann, Ruiz-Sanchez & Rojas-Soto, 2016). Likewise, species turnover for animal species are predicted to be high in the northern of Mexico (Peterson et al., 2002). Ecological niche models in the Mexican biota have been used extensively to predict future range shifts and species turnover, largely based on species occurrence data (e.g., Cantarello et al., 2011; Gómez-Mendoza & Arriaga, 2007; Ramírez-Amezcua et al., 2016). However, adding genetic data accounting for the role of gene flow and local adaptation across spatial scales would improve the prediction of such models (Fordham, Brook, Moritz & Nogués-Bravo, 2014). Moreover, recent approaches advocate for the integration of historical and contemporary habitat connectivity data into species distribution modelling to significantly increase the spatial accuracy of species distribution models (Scoble & Lowe, 2010).
On the other hand, forest genetic resources in Mexico are considered of fundamental importance for their economic, ecological, and social value (Food and Agriculture Organization of the United Nations [FAO], 2012). Landscape genomic-wide studies in Mexican forest tree species can reveal how environmental variables shape local and large-scale spatial variation of important phenotypic traits related to survival and growth (Alberto et al., 2013). Spatial information on genomic variation would better assist landscape management guidelines to delineate seed collection zones, management units, and mitigation measures like assisted migrations (Castellanos-Acuña, Lindig-Cisneros & Sáenz-Romero, 2015).
Recommendations & Conclusions
There are aspects to consider in future landscape genetics studies. First, reliable evaluations of spatial genetic structure require an accurate sampling design, which entails matching the temporal and spatial scale of the studied biological process driving genetic structure (Anderson et al., 2010). Equally important is the selection of the numbers of populations, sampled individuals, and molecular markers (Hall & Beissinger, 2014). Moreover, the choice of population- over individual-based sampling and analyses should not be trivial. For species that have continuous distributions and long distance dispersal, a population sampling approach of aggregated spatial samples with evident missing sites of occurrence may not capture the underlying pattern of genetic structure (Landguth & Schwartz, 2014; Schwartz & McKelvey, 2009). Most of the reviewed studies here did not report how and why sampled locations were selected, and neither did they justify the choice of population-over individual-based sampling/analysis or vice versa. A practical way to inform sampling design, including the performance of statistical methods, is the implementation of computer landscape genetic simulations (Oyler-McCance, Fedy & Landguth, 2013). For instance, species-specific simulations with model parameters focused on the life history traits of the selected species and its distribution in the landscape allowed to evaluate the most appropriate sampling scheme in forest tree species of the TDF in the Bajio Region in Mexico (Rico, 2017).
A second aspect to consider is the occurrence of IBD as it can significantly overestimate the identification of spatial genetic structure (Schwartz & McKelvey, 2009). Most of the studies evaluated here, which performed Bayesian and multivariate clustering methods (e.g., Structure, PCA), found signs of genetic structure (k> 3 clusters), but they also reported a significant pattern of IBD. Hence, it is not clear how much of the genetic structure found could be the result of biased inferences due to IBD. Methodological recommendations have been suggested to account for IBD when testing for hierarchical genetic structure (Meirmans, 2012), and landscape effects on gene flow (van Strien, Holderegger & Van Heck, 2014).
Lastly, accounting for historical effects on gene flow before testing for contemporary habitat connectivity is of particular relevance for many species ocurring in biogeographic complex regions in Mexico (e.g. TVB). For example, the study by Dyer et al. (2010) effectively demonstrated the importance of taking into account the phylogeographic pattern of the species for testing landscape genetic hypotheses, as failing to do so would lead to incorrect inferences of current landscape features on gene flow. Other approaches have been suggested by Epps & Keyghobadi (2015).
In conclusion, the diversity of questions that can be resolved by expliclitly testing the landscape context on gene flow is extensive. As shown here, the total number of species studied in Mexico within this framework is still few but which numbers are increasing. Because antropogenic activities are drastically transforming the natural world, we need to understand how such changes affect species persistence. Landscape genetics offers an interdisciplinary approach to better address such a complex conservation challenge.











 text new page (beta)
text new page (beta)