INTRODUCTION
Heavy metals are chemical elements with a density of at least five times that of water, a high atomic number, and are toxic and poisonous at low concentrations (Alloway 2012, Poschenrieder and Barceló 2015). Due to their abundance and toxicity, the most studied are Hg, As and Pb; to a lesser extent, Cr (Covarrubias et al. 2015).
The main problems associated with environmental pollution by heavy metals are bioaccumulation in the food chain and environmental persistence (Zahoor and Rehman 2009). Most heavy metals are released into water bodies because of anthropogenic activities such as wastewater discharges, industrial wastes, agricultural wastes, and mining (Yang et al. 2009).
Unlike organic pollutants, biological, chemical, or physical processes cannot permanently remove metals (Volke-Sepúlveda et al. 2005). Depending on their nature, biological methods, known as bioremediation techniques, can be used to remove them. In these cases, the metabolic potential of microorganisms (bacteria and fungi) and plants is used to clean contaminated ecosystems by mobilizing or immobilizing heavy metals (Lovley and Coates 1997, Gadd 2010). Bacteria are among the most widely used microorganisms in bioremediation processes (Schippers and Sand 1999) due to their specific genetic mechanisms of tolerance and interaction with various elements (Silver and Misra 1988, Mindlin et al. 2001).
For some bacterial species, low concentrations of metals such as Ni, Fe, Cu, and Zn are essential for their metabolism. In contrast, Hg, Ag, Cd, and Pb are toxic to most microorganisms, even at low concentrations, as they are not biologically active (Hughes and Poole 1989). Bacterial species with the ability to survive in the presence of heavy metals are of great interest for use in bioremediation processes.
Heavy metal tolerance studies have also been conducted with actinobacteria, as described by Yang et al. (2009), who isolated Intrasporangium sp. Q5-1 from the soil of a manganese-chrome mine, finding that it is highly resistant to Cr6+ and efficiently removes this metal under aerobic conditions. El-Baz et al. (2015)isolated and identified Streptomyces sp. BN2, Amycolatopsis tucumanensis and Amycolatopsis sp. GT6, GT15, and GT39 from mining areas in the Marrakech region of Morocco. The Streptomyces sp. BN2 strain was tolerant to Pb2+ at a maximum concentration of 0.55 mg/mL. A. tucumanensis tolerated Cu2+ at a maximum concentration of 0.08 mg/mL, while Amycolatopsis sp. GT6, GT15, and GT39 tolerated Pb2+, Cu2+, Cr6+, and Zn2+ at maximum concentrations of 0.25, 10.10, 0.15 and 0.10 mg/mL respectively. On the other hand, Bueno et al. (2008) tested the bioaccumulation of Pb2+, Cr3+, and Cu2+ in Rhodococcus opacus at pH 5 and 25 ºC. They concluded that the species bioaccumulated 94.3, 72.9, and 32.2 mg/mL of Pb2+, Cr3+, and Cu2+, respectively.
Several microorganisms with the ability to tolerate heavy metals have been isolated in poorly studied ecosystems, such as saline environments (Haefeli et al. 1984, Duxbury 1986, Basu et al. 1997, Choudhury and Kumar 1998, Castro-Silva et al. 2003, Otth et al. 2005, Lima-Bittencourt et al. 2007). In these sites, predominantly halophilic bacterial species develop, requiring high concentrations of NaCl to grow and survive (Oren 2006).
According to Ramirez et al. (2005), it is possible to distinguish weak halophiles, with optimum growth at around 3% NaCl; moderate halophiles, with optimum growth in a range of 3 to 15% salt; extreme halophiles, with optimum growth at 25% NaCl. Likewise, non-halophilic microorganisms capable of growing both in the absence and presence of salt are called halotolerant; those that tolerate up to 15% NaCl are considered extreme halotolerant.
Due to their physiological activity, it has been shown that halophilic and halotolerant bacteria can thrive in high concentrations of metals even though their habitat does not necessarily contain them (Al-Mailem et al. 2018). Passive transport is a process free of bacterial metabolism and unrelated to an expenditure of energy by which metal biosorption occurs. This happens when binding sites on the bacterial surface cause metal cations to become attached.
On the other hand, a process that does involve energy expenditure and may be included in metabolic pathways is active transport. This occurs when metals enter cells directly and, in both cases, is considered bioaccumulation (Lyer et al. 2004). In addition, there is a direct relationship between salinity and metal uptake. This relationship shows that metal cations play a role in cytoplasmic osmolality or are adsorbed by bacteria as a tolerance strategy (Dell’Amico et al. 2005).
Several extreme saline environments have been identified in Mexico, such as the ancient Lake Texcoco. Halophilic heavy metal-tolerant bacteria may be present at this site and may have the potential for bioremediation. However, bacteria from this region are not known to have this potential; therefore, the present study aimed to isolate and identify halophilic bacteria tolerant to heavy metals from soil samples from the ancient lake of Texcoco, Mexico.
MATERIALS AND METHODS
Sampling and physical and chemical characterization of samples
A simple random surface sampling in triplicate soil and saline-sodic sediment was carried out in seven areas of the former Lake Texcoco, Mexico (Latitude: 19.472601, Longitude: -98.945665, altitude: 2236 m). The physical and chemical parameters of NaCl percentage and pH were determined for all samples. For this purpose, dry soil (1 g) of each sample was placed in a test tube, and 9 mL of distilled water was aggregated. The mixture was vortexed for 10 min and filtered overnight through Whatman No. 2 filter paper. Determination of % NaCl and pH in the filtrate was carried out using a refractometer (Hanna, ref. HI931100) and a potentiometer (Hanna, ref. HI98128) respectively.
Culture medium for halophilic bacteria
A culture medium for halophilic bacteria (HM) was prepared (Ventosa et al. 1982): 10 g/L yeast extract, 5 g/L protein peptone, 1 g/L glucose, 18 g/L bacteriological agar, and 10% NaCl solution. The pH was adjusted to 7.2 ± 0.2. The medium was autoclaved at 121 ºC for 15 min, cooled to 50 ºC, and poured into Petri dishes. Plates were maintained at 37 ºC for 24 hours to check sterility.
Isolation and purification of halophilic bacteria
From each sample, 1 g of soil was taken, placed in a test tube, and 9 mL of 10% (w/v) NaCl solution was added. The mixture was vortexed for 30 s. Aliquots of 200 µL of each suspension were inoculated into the HM culture medium and incubated for 10 days at 37 ºC. At the end of this period, the strains were isolated and purified by reseeding in the same medium until a single strain was obtained in each Petri dish, confirming the purification by the uniformity of cell morphology.
Morphological characterization of the strains
The macroscopic morphology of the purified bacterial strains was described according to the following characteristics: size, color, shape, texture, and presence of aerial or vegetative mycelium. The microscopic morphology of the strains was determined by the response to Gram staining, shape, clustering, and the presence of filamentous cells and branching.
Physiological characterization of halophilic bacteria
NaCl concentration and pH were measured for optimal growth for physiological characterization of the isolated strains. The optimal NaCl concentration was determined by seeding each strain in HM medium adjusted to different concentrations: 0, 1, 3, 5, 5, 10, 15, 15, 20, 25 and 30% (w/v) at pH 7.2 ± 0.2. Seeding was performed by streaking; plates were incubated at 37 ºC, and colony growth was observed for 10 days. The optimum NaCl concentration for growth was that at which NaCl was most abundant.
The optimum pH value for the growth of the purified strains was determined using HM culture medium adjusted to the corresponding concentration of the NaCl concentration previously determined for optimum growth. The pH was adjusted to 6, 7, 8, 8, 9, 10, 11, and 12. Each strain was streak inoculated onto HM medium and incubated at 37 ºC for 10 days, during which growth was observed. The optimum pH for the growth of each strain was determined as the value at which colony growth was most abundant.
Tolerance of bacterial strains to heavy metals
The tolerance of the strains to each of the metals was determined as follows. The strains were inoculated into flasks with HM medium at 10% NaCl and pH 8.0 ± 0.2, supplemented with the corresponding metal in the following chemical species and concentrations As3+ (0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0 and 12.0) mmol/L; Cr6+ (0.0, 100 0, 200.0, 300. 0, 400.0, 500.0, 600.0, 700.0, 800.0, 900.0, 1 000.0, 1 100.0 and 1 200.0) mmol/L; Hg2+ (0. 0, 0.005, 0.01, 0.015, 0.02, 0.025, 0.03, 0.35, 0. 04, 0.45, 0.05, 0.55, 0.06, 0.65, 0.07, 0.75, 0. 08, 0.85, 0.09, 0.95 and 0.1) mmol/L; Pb2+ (0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0 and 8.0) mmol/L. The flasks were kept on an orbital shaker at 150 rpm and 37 ºC for 15 days to determine the highest metal concentration at which the strains were able to grow. Heavy metal tolerant strains were defined as bacteria able to reproduce after incubation for 15 days in the presence of the above metal concentrations.
Genetic identification of the heavy metal-tolerant strains
Strains that tolerated the highest concentrations of one or more metals were genetically identified by 16S rRNA gene sequence analysis. For this purpose, strains were seeded in HM medium at 10% NaCl and pH 8.0 ± 0.2 and incubated at 37 ºC and 45 x g for one week. The biomass was then collected by centrifugation at 3000 x g for 10 minutes. According to the manufacturer’s instructions, bacterial DNA extraction was performed with the Wizard® genomic DNA purification kit (Promega A1120). The presence and integrity of DNA were checked by 1% agarose gel electrophoresis (PROMEGA V3121), stained with ethidium bromide (SIGMA 46065), with a run time of 45 minutes, 120 V.
Subsequently, the extracted DNA was used as a template for 16S rRNA gene amplification by polymerase chain reaction (PCR). Taq DNA Polymerase (MyTaq, Bioline BIO21105) and the following universal primers were used: 27F: 5’ AGAGTTTGATCMTGGCTCAG 3’; 518F: 5’ CCAGCAGCCGCGGTAATACG 3’; 1492R: 5’ TACGGYTACCTTGTTGTTACGACTT 3’; 800R: 5’ TACCAGGGTATC TAATCC 3’. Thermal cycling conditions were pre-denaturation for 5 min at 94 ºC; followed by 34 cycles of denaturation for 30 s at 94 ºC, annealing for 20 s at 52 ºC and extension for 90 s at 72 ºC; and final extension for 7 min at 72 ºC. The amplified fragments were visualized by 1% agarose gel electrophoresis (Promega V3121) stained with ethidium bromide (Sigma 46065) with a 1 kb molecular weight marker (Thermo Scientific SM0313) and run time of 45 min, 120 V.
Amplification products were purified using the Amicon Ultra centrifugal filter unit (Millipore® UFC901008) and sent to the Macrogen DNA sequencing service (Maryland, USA). The electropherograms of each sequence were checked using the ChromasPro v1.5 program (Technelysium, Pty). The ends of each sequence, corresponding to the primers used, were trimmed. Once clean sequences were obtained, they were saved in FASTA format.
The sequences in FASTA format were entered into the BioEdit v7.0.9 program (Hall 1999) and assembled using the contig assembly program (CAP) (Huan 1992) with a minimum overlap of 20 bases and 80% coincidence. The consensus sequences obtained were compared with sequences validated in the GenBank database (National Center for Biotechnology Information, NCBI) using the Nucleotide Basic Local Alignment Search Tool (BLASTn) program (Altschul et al. 1990) and the public database EZ-Biocloud (Yoon et al. 2017).
Phylogenetic analysis
The consensus sequences of the isolated strains and 16S rRNA gene sequences retrieved from the GenBank database were compared for phylogenetic analysis. The alignment was generated in MEGA (Molecular Evolutionary Genetics Analysis) v7.0.21 (Kumar et al. 2016), which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega. The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI. This alignment was used to estimate the best surrogate model to construct the phylogenetic tree. The phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei 1987). Branch support was evaluated with a bootstrap value of 1000 replicates (Felsenstein 1985).
Determination of minimum inhibitory concentration (MIC)
The MIC of identified strains was determined to find the exact concentration inhibiting their growth. Strains were cultured in HM medium with 10% NaCl at pH 8.0 ± 0.2, supplemented with the respective metal at the following concentrations: As3+, 3.0-13.0 ± 0.25 mmol/L; Cr6+, 300.0-1300.0 ± 25 mmol/L, Hg2+, 0.0-0.1 ± 0.025 mmol/L; and Pb2+, 3.0-10.0 ± 0.5 mmol/L. Bacterial strains were incubated for 15 days to determine the concentration at which they could no longer grow.
Growth curve and morphological changes in the presence of heavy metals
Two flasks with HM culture medium at 10% NaCl and pH 8.0 ± 0.2 were set up for each strain identified, and one flask was supplemented with the metal’s maximum tolerable concentration (MTC). The strain inoculum in each flask was adjusted to McFarland standard No. 2. The flasks were maintained on an orbital shaking platform at 45 x g and 37 ºC. A growth curve was plotted for each flask to evaluate its behavior under normal conditions (without metal) and in the presence of the tolerated metal. Bacterial growth was determined by turbidimetry in aliquots taken from the flask at different intervals. The optical density was measured at 660 nm.
Two Petri dishes were prepared in duplicate with HM medium at 10% NaCl and pH 8.0 ± 0.2 to complement the test. One plate was added to the MTC of the metal. The corresponding strains were seeded and incubated at 37 ºC for 15 days. At the end of this period, the morphological characteristics of colony growth were macroscopically examined, including size, color, form, texture, and presence of aerial or vegetative mycelium. The comparison was made between colony growth in normal conditions (no metal) and the presence of the tolerated metal.
RESULTS
Sample characteristics
Twenty-one samples were collected, 15 of soil and six of sediment. Sampling was carried out in dry and rainy seasons and included seven areas of the surface of former Lake Texcoco (zones 1-7). The physical and chemical characterization of the samples is detailed in table I. The percentage of NaCl ranged between 3.61 and 14.26%, and the pH values were between 9.66 and 12.01.
TABLE I PHYSICAL AND CHEMICAL CHARACTERISTICS OF THE COLLECTED SAMPLES.
| Sampling area number | Percentage of NaCl | pH | Characteristics of the sampling area | UTM* Zone 14Q |
| Zone 1 | 3.61 | 11.61 | Wet-muddy | X:0500795 Y:2152079 |
| Zone 2 | 6.18 | 10.55 | Dry grassland | X:0502962 Y:2153770 |
| Zone 3 | 4.08 | 9.66 | Dry soil | X:0502721 Y2153770 |
| Zone 4 | 5.26 | 11.60 | Dry soil | X0502714 Y:2155311 |
| Zone 5 | 14.26 | 12.01 | Waterway | X:0501405 Y:2155656 |
| Zone 6 | 8.68 | 11.65 | Waterway | X:0500666 Y:2157378 |
| Zone 7 | 9.66 | 11.72 | Wet-muddy | X:0500697 Y:2157372 |
*Universal Transverse Mercator
Isolated strains
A total of 75 bacterial strains were isolated; all showed growth in HM medium at 10% NaCl and pH 7.2 ± 0.2. One of the main macroscopic morphological characteristics was coloration. The strains identified were yellow, orange, or white. They were circular or irregular in shape and had smooth or wavy margins, a smooth texture, and a rough or mucoid consistency; some were shiny, and others were opaque. As for the microscopic characteristics, Gram-positive and Gram-negative forms and bacillary and coccoid forms were observed. Specifically, strains TX3SA-2MHG1 and TX3SA-4MHG1 were Gram-positive cocci; strain TXO4B-1SG9 showed Gram-negative bacillary morphology. Strains TXV10-3SG5 and TXV7-8SG2 were Gram-positive filamentous bacteria with associated coccoidal structures.
Physiological characterization of halophilic bacteria
All strains required a minimum concentration of at least 3% NaCl to grow; none grew in the absence of salt, and, therefore, all were classified as halophilic. The strains could grow to a maximum concentration of 15% NaCl, classified as moderate halophilic. All strains grew to a maximum pH value of 10, started growth at pH 6-7, and were therefore classified as alkali-tolerant. Optimal growth of most strains was found at 10% NaCl and pH 8; therefore, most strains were classified as moderate halophilic and alkali-tolerant bacteria.
Tolerance of bacterial strains to heavy metals
In the case of As3+ and Pb2+, of the 75 strains evaluated, at least one tolerated each of the concentrations tested. For Cr6+, 69 strains tolerated at least one of the concentrations tested, while only 39 did so for Hg2+ (Table II). Strains were identified that tolerated high concentrations of the metals. The most toxic metal of all was Hg2+, as the lowest number of strains tolerated it, and its MIC was much lower than the other metals (Table II).
TABLE II DISTRIBUTION OF THE ISOLATED BACTERIAL STRAINS, ACCORDING TO THE MAXIMUM TOLERABLE CONCENTRATION (mmol/L) OF THE HEAVY METALS EVALUATED.
| Metal | Concentration (mmol/L) and number of strains | ||||||||||
| As3+ | Concentration | 1.0 | 3.0 | 4.0 | 5.0 | 6.0 | 8.0 | 27.0 | |||
| No. of strains | 7 | 35 | 22 | 4 | 2 | 4 | 1 | ||||
| Cr6+ | Concentration | 0 | 100 | 200 | 300 | 400 | 500 | 600 | 700 | 900 | 1200 |
| No. of strains | 6 | 19 | 9 | 3 | 16 | 1 | 2 | 3 | 15 | 1 | |
| Hg2+ | Concentration | 0.000 | 0.005 | 0.010 | 0.015 | 0.020 | 0.060 | 0.080 | |||
| No. of strains | 36 | 11 | 15 | 2 | 9 | 1 | 1 | ||||
| Pb2+ | Concentration | 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | |||
| No. of strains | 6 | 3 | 8 | 48 | 8 | 1 | 1 | ||||
Identification of strains with higher tolerance to heavy metals
Strains that showed tolerance to the highest concentrations of one or more of the metals evaluated were identified by comparative analysis of the 16S rRNA gene sequence. The size of the sequences ranged from 1222 to 1492 base pairs. Three bacterial genera were detected: Salinicococcus spp. strains TX3SA-2MHG1 and TX3SA-4MHG1, Halomonas sp. strain TXO4B-1SG9, and Nocardiopsis spp strains TXV10-3SG5 and TXV7-8SG2 (Table III). The sequences were deposited in NCBI GenBank with accession numbers KY10903-4 and KY10906-9 (Table III).
TABLE III HALOPHILIC HEAVY METAL TOLERANT BACTERIA IDENTIFIED BY 16S rRNA GENE SEQUENCE ANALYSIS.
| No. Strain | Fragment size (bp) | Genbank Access | BLAST | Ez Biocloud | ||||
| Most similar species | Identity percentage | Cover percentage | Most similar species | Identity percentage | Cover percentage | |||
| 1 TX3SA-2MHG1 | 1420 | KY710903 | Salinicoccus roseus | 98.10 | 100 | Salinicoccus roseus | 98.28 | 95.2 |
| 2 TX3SA-4MHG1 | 1485 | KY710904 | Salinicoccus roseus | 98.52 | 100 | Salinicoccus roseus | 98.97 | 99.7 |
| 3 TXO4B-1SG9 | 1440 | KY710906 | Halomonas pantelleriensis | 99.03 | 100 | Halomonas pantelleriensis | 99.23 | 98.4 |
| 4 TXO7B-1SG12 | 1492 | KY710907 | Nocardiopsis xinjiangensis | 98.86 | 99 | Nocardiopsis xinjiangensis | 99.11 | 100.0 |
| 5 TXV10-3SG5 | 1424 | KY710908 | Nocardiopsis xinjiangensis | 98.74 | 100 | Nocardiopsis xinjiangensis | 99.23 | 97.9 |
| 6 TXV7-8SG2 | 1222 | KY710909 | Nocardipsis salina | 99.18 | 99 | Nocardipsis salina | 98.85 | 83.9 |
No.: strain number; bp: base pairs
Phylogenetic analysis
A phylogenetic tree was constructed based on the 16S rRNA gene sequences of the isolated strains and the sequences obtained from GenBank for each genus. The tree shows the isolated strains forming three groups (Fig. 1). The first corresponds to the strain identified as Halomonas spp. (TXO4B-1SG9), where a close evolutionary relationship with the species H. pantelleriensis is observed. The second was formed by the strains identified as Salinicococcus spp. This shows that strain TX3SA-4MHG1 has a strong evolutionary relationship with the species S. roseus, while strain TX3SA -2MHG1 has a stronger evolutionary relationship with the species S. iranensis. The third group corresponds to the strains identified as Nocardiopsis spp., where it is observed that both strain TXV7-8SG2 and strain TXV10-3SG5 have a stronger evolutionary relationship with the species N. salina.

Fig. 1 Phylogenetic tree based on 16S rRNA gene sequences, showing the phylogenetic relationships of halophilic metal-tolerant strains. Constructed using the neighbor joining method. The boostrap value of 1000 replicates was used. The scale bar represents the number of substitutions per site. GenBank accession numbers are indicated in parentheses.
Specifically, strain TXV10-3SG5 had been previously identified as N. xinjiangensis by sequence comparison with BLAST and EZ-Biocloud programs. Similarly, strain TXO7B-1SG12, previously identified as N. xinjiangensis, is evolutionarily distant from this species, being more closely related to N. nikkonensis. Phylogenetic analysis confirmed the identification of the genera of the isolated strains (Fig. 1).
MICs of heavy metals in the identified strains
Regarding minimum inhibitory concentrations, the strain Nocardiopsis sp. TXV7-8SG2 tolerated three metals (As3+, Cr6+, and Pb2+) at their highest concentration. Although it also tolerated a high concentration of Hg2+, the strain resistant to the highest concentration of this metal was Salinicoccus sp. TX3SA-4MHG1. The MICs of the metals evaluated in the identified strains are detailed in table IV.
TABLE IV CLASSIFICATION, TOLERANCE, AND MINIMUM INHIBITORY CONCENTRATION (MIC) OF BACTERIAL STRAINS THAT PROVED ABLE TO GROW IN THE PRESENCE OF HIGHER HEAVY METAL CONCENTRATIONS
| Strain | Range of % NaCl for growing/ Classification | Range of pH for growing/Classification | Metal | Tolerance (mmol/L) | MIC (mmol/L) |
| Nocardiopsis sp. TXV7-8SG2 | 3-15 moderate halophilic | 7-10 alkalotolerant | As3+ | 27.00 | 27.25 |
| Cr6+ | 1 200 | 1 250 | |||
| Hg2+ | 0.060 | 0.065 | |||
| Pb2+ | 7.00 | 7.75 | |||
| Nocardiopsis sp. TXO7B-1SG12 | 5-15 moderate halophilic | 6-10 alkalotolerant | As3+ | 8.00 | 8.20 |
| Nocardiopsis sp. TXV10-3SG5 | 3-15 moderate halophilic | 6-10 alkalotolerant | As3+ | 9.00 | 9.75 |
| Salinicoccus sp. TX3SA-2MHG1 | 5-15 moderate halophilic | 7-10 alkalotolerant | Hg2+ | 0.06 | 0.06 |
| Salinicoccus sp. TX3SA-4MHG1 | 5-15 moderate halophilic | 7-10 alkalotolerant | Hg2+ | 0.08 | 0.10 |
| Halomonas sp. TXO4B-1SG9 | 5-15 moderate halophilic | 6-10 alkalotolerant | Pb2+ | 6.00 | 7.00 |
Effect of heavy metals on bacterial growth
As a result of exposure of the strains to the MTC of individual metals, growth curves were obtained (Fig. 2), showing the effect of the tolerated metal on the corresponding strain. These curves reveal that metal presence extended the latency period in some cases, lengthening the time required to reach maximum growth, while in other cases, metal presence did not extend the latency period but prevented maximum growth attainment as compared to growth in the absence of metals.
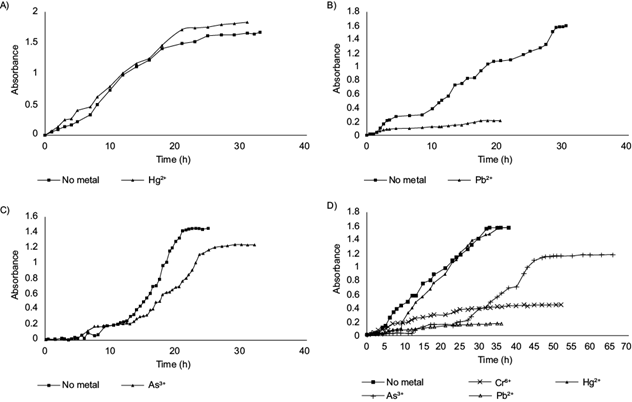
Fig. 2 Growth curves of bacterial strains in the presence/absence of the tolerated heavy metal. A) Salinicoccus sp. TX3SA-2MHG1 growth, without metal and with 0.08 mmol/L (Hg2+). B) Halomonas sp. TXO4B-1SG9 growth, without metal and with 6.00 mmol/L (Pb2+). C) Nocardiopsis sp. TXO7B-1SG12 growth, without metal and with 9.00 mmol/L (As3+). D) Nocardiopsis sp. TXV7-8SG2 growth, without metal and with 27.00 mmol/L (As3+), 1200 mmol/L (Cr6+), 0.07 mmol/L (Hg2+), and 7.00 mmol/L (Pb2+).
Likewise, figure 3 shows that the presence of metals induced morphological changes in some heavy metal tolerant strains. The strain Salinicoccus sp. TX3SA-2MHG1, in the presence of Hg2+, presented smaller pale orange colonies; the colonies had an elevated center and developed confluent, although it had the same texture and maintained the absence of aerial mycelium. Halomonas sp. TXO4B-1SGP, in the presence of Pb2+, presented a slightly smaller, dark brown colony; it maintained the same texture, and there was an absence of aerial mycelium. Nocardiopsis sp. TXV10-3SG5, in the presence of As3+, did not show changes in size or color. Rounded edges were observed, and the same texture and the presence of aerial mycelium were maintained.
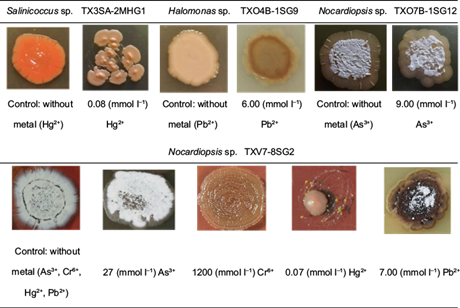
Fig. 3 Macroscopic images of growth and morphology in control strains (without heavy metals) as compared to strains growing in the presence of heavy metals.
On the other hand, Nocardiopsis sp. XV7-8SG2, in the presence of As3+, maintained the same morphological characteristics as its growth without metal (control). In the presence of Cr6+, this species did not change its size, but the color changed to light brown; the shape and texture were the same, and the absence of aerial mycelium was observed. The same strain in the presence of Hg2+ notably decreased in size, changed to a light orange color, and presented a semi-rounded shape, smooth texture, and absence of aerial mycelium. Finally, this strain in the presence of Pb2+ had the most noticeable changes; the colony size decreased, the color changed to dark brown with some black areas, rounded edges were observed, the smooth texture was maintained, and the decrease of aerial mycelium was observed (Fig. 3).
DISCUSSION
Heavy metal contamination is a severe environmental problem since physical and chemical remediation requires high costs, and complete removal is difficult (Prabhakaran et al. 2016). Bioremediation mediated by metal-resistant bacteria seems a suitable alternative.
Our research found that the bacteria evaluated showed resistance to several metals. And it is essential to mention that the literature indicates that bacteria isolated from saline or hypersaline environments are valuable models for investigating the molecular basis of metal tolerance (Xu et al. 2013). Numerous transport systems (metal-specific efflux pumps), transcriptional regulatory factors, and reducing enzymes that could mediate metal tolerance have been found in bacterial genomics-related research (Zhou et al. 2014). In some cases, genes encoding and conferring multi-resistance to metals such as Ni, Cu, Cr, As, Zn, Co, and Cd have been detected, for example, in the genome of Marinobacter manganoxydans MnI7-9 (Zhou et al. 2014).
In the same context, plasmid operons such ars, cad, or mer confer resistance to As, Cd, or Hg, which, through enzyme expression, act as metal detoxification, uptake, or transport agents (Sri et al. 2012). For example, metal resistance operons have been detected in halophilic bacteria such as Halobacterium sp. (Ng et al. 2000). It is thought that when a microorganism is in the presence of metals, it increases transcriptional action, which aims to reduce metal ingress and decrease lethal toxicity; this response could be transient as when the microorganisms manage to adapt to the metals in the surrounding environment, the transcript levels of the genes encoding the response return to typical values (Srivastava and Kowshik 2013).
In the present study, halophilic bacteria showed growth at concentrations of 3-10% NaCl (w/v), pH values 6-10, and 37 ºC. Therefore, microorganisms exhibiting these two physiological properties are expected to be able to grow in contaminated environments within these NaCl and pH ranges. Among these bacteria is the genus Streptomyces sp. Amoozegar et al. (2008) reported that this genus, specifically S. roseus, tolerates tellurium (Te) at a MIC of 12 mmol/L under 10% NaCl, pH 7-8, and 34-38 ºC. This strain eliminated 75% of the metal in the medium in 72 h. Moreno et al. (2012) have also identified this species and its tolerance to Cd, Zn, Ni, Fe, Cu, and Co. In our study, strain TX3SA-2MHG1, genetically matched to S. roseus, showed the highest tolerance to Hg2+ with a MIC of 0.1 mmol/L.
Enzymatic processes can achieve the resistance of microorganisms to Hg. These processes are mercuric reductase (MerA) and organomercurial lyase involved in cellular detoxification (MerB). The latter cleaves the organic bonds of Hg, such as methylmercury and phenylmercury, causing the release of inorganic Hg2+; the MerA enzyme acts, capturing the Hg2+ and reducing it to volatile Hg0. Although the enzymatic action is the leading cause of Hg detoxification, there are also genes involved in this process that confer resistance to the bacteria, for example, the one coding for the MerP protein, which eliminates the Hg2+ captured in the periplasmic space with the help of the Hg2+ transporter in halophiles (Boyd and Barkay 2012, Zhou et al. 2014).
On the other hand, VanEngelen et al. (2008)identified the genus Halomonas sp., which showed tolerance and reduced Cr6+ and Fe3+ under alkaline conditions. Strain TXO4B-1SG9 isolated in our study belongs to this genus and showed the highest tolerance to Pb2+ (MIC = 7.0 mmol/L) and a MIC for Cr6+ of 900 mmol/L, which is not the most tolerated concentration, but still high.
The genus Nocardiopsis sp. (strains TXO7B-1SG12 and TXV10-3SG5) had a high tolerance to As3+ (MIC = 9.75 mmol/L). Likewise, strain TXV7-8SG2 recorded the highest tolerance to three of the metals evaluated, with MICs of (27.25, 1250.00, and 7.75) mmol/L for As3+, Cr6+ and Pb2+, respectively. It also showed substantial resistance to Hg2+ (MIC = 0.075 mmol/L). The present study reports tolerance to the metals evaluated here in these strains.
As mentioned above, one reaction mechanism to metals is enzymatic, which attempts to reduce metal species by transforming them into less harmful ones. The primary example is the biological reduction of Cr6+, which is enzymatically reduced to Cr3+ (a fewer toxic species that can participate in biochemical processes) under haloalkaline conditions. This mechanism involves the action of chromate reductases, located in the cell membrane, specifically for moderate halophilic bacteria, the NADH-dependent reductase (Focardi et al. 2012). This has been demonstrated in strains of Amphibacillus and Halomonas species isolated from uncontaminated and Cr6+ contaminated sites, respectively (Mabrouk et al. 2014). In our experiment, the strain TXV7-8SG2 may have used this enzymatic process as a mechanism of tolerance/resistance to Cr6+ by tolerating up to 1200 mmol/L since precise information is not available as it is a species exposed for the first time to these metal concentrations.
Bacterial growth in the presence of As3+ showed no apparent changes in morphology since the strains developed the same shape and color, perhaps indicating that element does not affect their growth. Colony growth in the presence of Cr6+, Pb2+, and Hg2+ induced changes. The strains acquired a darker color, probably due to the reduction of Cr6+ to Cr3+ and the formation of PbS and HgS.
Finally, 75 halophilic strains isolated from ancient Lake Texcoco showed tolerance to As3+ and Pb2+ at concentrations of 1.0-27.0 mmol/L and 1.0- 7.0 mmol/L, respectively. Of these strains, 69 were resistant to Cr6+ at 100-1200 mmol/L and 39 to Hg2+ at 0.005-0.80 mmol/L concentrations Cr6+, Hg2+, and Pb2+ induced morphological changes in the growth of these strains, whereas As3+ had no such effect.











 text new page (beta)
text new page (beta)


