INTRODUCTION
The anaerobic ammonium-oxidizing (Anammox) is a bacterial metabolic pathway of the nitrogen cycle in which ammonium is directly oxidized to dinitrogen (N2) in an anoxic environment with nitrite as a final electron acceptor. The overall catabolic reaction is (Jetten et al. 1999):
Anammox microorganisms constitute a distinct phylogenetic group of bacteria, predominantly within the order of the Planctomycetales (Kartal et al. 2013). They are especially characterized by their Anammoxosome, a cell compartment where Anammox metabolism takes place, and by their very slow rate of division (Fuers et al. 2006, Fernández et al. 2014).
Despite this, in the oxygen-limited zones, the role of Anammox bacteria in the worldwide production of N2 is today known to be important. Recent estimates suggest that this metabolism may be responsible for 30-50 % of N2 production in oceans (Devol 2003). The past decade has seen growing interest in those bacteria due to their capacity to eliminate ammonium with a very low consumption of biomass. Moreover, as part of a process to treat nitrogen pollution, a strategy based on Anammox bacteria would allow shortening the process from two steps to one.
Anammox bacteria were first observed in wastewater treatment plants (Ren et al. 2020) but were later spotted in a wide range of natural environments (Nie et al. 2019). They were found in oceanic regions and anoxic basin (Kuypers et al. 2003), as well as deep-sea hydrothermal vents (Byrne et al. 2008). The Anammox process was also observed in freshwater zones, in hot springs (Jaeschke et al. 2009), estuaries and continental sediments (Trimmer et al. 2003, Jaeschke et al. 2010). More recently, some studies stated presence of Anammox bacteria in different types of terrestrial ecosystems, in lakeshores, wetlands and agricultural soils (Humbert et al. 2010, 2012, Wang and Gu 2013, Shen et al. 2013, Pajares and Bohannan 2016). Other studies aimed to determine if Anammox bacteria were able to adapt to extreme environments. The inhibitory effects of several culture conditions in reactors were demonstrated (Jin et al. 2012, Yu and Jin 2012) and it was particularly stated that high pH (> 9) induced an important decrease of Anammox activity. However, Anammox bacteria were also discovered in very high-temperature water containing metal sulfides (Russ et al. 2013), marine hypersaline basins (Borin et al. 2013), high-temperature petroleum reservoirs (Li et al. 2010), and acidic agricultural soils (pH < 4) (Shen et al. 2013).
The former Lake Texcoco is an ancient salty lake located in the Valley of Mexico, more than 2000 masl. This closed lake without current water outfall was drained to counter major problems of chronic flooding in nearby Mexico City. Its soils are saline and sodic, with a pH between 9.8 and 10.4, electrical conductivities in saturation extracts between 22 and 150 dS/m and a large percentage of exchangeable sodium (60-80) (Luna-Guido et al. 2000, Rojas-Oropeza et al. 2010). This site is also characterized by the presence of a network of canals of wastewater and a deficit of sewage treatment (Gómez and Díaz 2011). Ammonium was also brought into the soil during irrigation projects which used wastewater to drain salts and fertilization attempts that used sewage sludge (Beltrán-Hernández et al. 1999). Due to the high pH of Texcoco soil, an important proportion of ammonia volatilizes in form of ammonia (du Plessis and Kroontje 1964, Vlek and Craswell 1981, Rojas-Oropeza et al. 2010). In addition, Rojas-Oropeza (2012) showed the presence of the Anammox functional gene hzoA in soils extracted from the former Lake Texcoco.
In this context, a better understanding of nitrogen cycles is of high interest. Therefore, in this study, our objective was to study the presence of the Anammox functional gene hzoA with its potential activity in extremophile soils of saline-sodic soils of the former Lake Texcoco. The hypothetic presence of Anammox microorganisms would be in relation to the anaerobic oxidation of ammonium. This will be done by analyzing the conversion rates of the different forms of nitrogen in soil samples of the former Lake Texcoco subjected to a helium atmosphere in a microcosm.
MATERIALS AND METHODS
Sampling sites
The study area (the former Lake Texcoco) is located in the Valley of Mexico at an altitude of 2210 masl, along the northeast part of Mexico City (Beltrán-Hernández et al. 1999). The superficial soil appears dry, brittle to the touch, and often contains cracks and salt crusts. The dominant vegetation on the site is saline grass (Distichlis spicata), which in the sampling area was < 15 cm high and sparsely distributed or absent due to the high salinity and sodicity. Six distinct sites were selected for sampling. In three of them, the salinity was considered as high (conductivity: 119.93 ± 13.9 dS/m, pH 10.0 ± 0.1), while in the three other sites it was lower (conductivity: 34.2 ± 2.8 dS/m, pH 9.1 ± 0.1). The 0-10 cm layer after removing the superficial salt crust (1 cm depth) was sampled 18 times and the soil of six plots was pooled so that three soil samples were obtained in each sampling site (n = 9 for each salinity) (February 2014). The soil samples were 2-mm sieved and characterized by triplicate. The physical and chemical properties of these soils are shown in table I: pH in a 1:2.5 soil-H2O suspension using a glass electrode; electrolytic conductivity in a saturated extract; water-holding capacity of soil samples saturated with water in a funnel overnight (Rhoades et al. 1989, Thomas 1996).
TABLE I PHYSICAL AND CHEMICAL CHARACTERISTICS OF SOILS SAMPLED AT THE FORMER LAKE TEXCOCO (FEBRUARY 2014).
| Soil sampled | pHH2O | ECs a DS/m | WHCb g/kg dry soil | Humidity % | Vegetation Description |
| Salinity 1 | 9.1 ± 0.1 | 34.2 ± 2.8 | 580 | 18.0 ± 1.6 | Abundant Distichlis spicata (grass height: 30 cm), little Eragrostis obtusiflora, little Chenopodium mexicana |
| Salinity 2 | 10.0 ± 0.1 | 119.93 ± 13.9 | 540 | 17.4 ± 0.2 | Little Distichlis spicata (grass height: 10 cm) |
aECs: electrolytic conductivity in the saturated extract; bWHC: water-holding capacity.
Direct DNA extraction of soil
Extraction of DNA from soil was done as described by Martin-Laurent et al. (2001). One milliliter of a solution containing 100 mM Tris (pH 8.0), 100 mM EDTA, 100 mM NaCl, 1 % (wt/vol) polyvinylpyrrolidone, and 2 % (wt/vol) sodium dodecyl sulphate was added to 250 mg soil in a 2 mL mini-bead-beater tube containing 0.5 and 0.1 g of 106 mm- and 2 mm-diameter glass beads, respectively. Samples were homogenized for 30 s at 1600 rpm in a minibead-beater cell disruptor (Mikro-Dismembrator S; B. Braun Biotech International). Samples were centrifuged at 14 000 × g at 4 ºC for 1 min. The collected supernatants were incubated with 5 M sodium acetate (1:10) on ice for 10 min and centrifuged at 14 000 × g for 5 min. After precipitation with one volume of ice-cold isopropanol, the nucleic acids were washed with 70 % ethanol and purified using a Sepharose 4B spin column. The quality and size of the soil DNAs were verified by electrophoresis on 1 % agarose gels. DNA was quantified using a BioPhotometer at 260 nm (Eppendorf, Hamburg, Germany). Three replicates were used for DNA evaluation of each soil sample.
hzoA gene amplification
The Anammox functional gene hzoA was amplified from 50 ng of DNA extracted from soil with a total volume of 25 µL by using 0.7 mL of the specific primers hzoA (9118-TATGGG TATGTC GATGGC TGA-9138 and 9583-CAACCA ACGATC CATACC GAA-9604) (designed in this work, based on Planctomycete KSU-1), and 2.5 U of Taq DNA polymerase (Appligene) under the following conditions: 5 min at 94 ºC, 1 min at 80 ºC (TaqPol); 1 cycle of 1 min at 94 ºC, 1 min at 55 ºC, and 1 min at 72 ºC; 10 cycles of 30 s at 94 ºC, 30 s at 55 ºC (with a decrease of 0.5 ºC per cycle), and 30 s at 72 ºC; and 30 cycles of 30 s at 94 ºC, 30 s at 52.5 ºC, and 30 s at 72 ºC, plus an additional cycle of 10 min at 72 ºC.
With the same conditions and after obtaining amplifications, these same reactions were used as template and amplifications were made with a reverse primer having at the 5’ end a GC clamp (9583-CGCCCGC CGCGCG CGGCGG GCGGGG CGGGGG CACGGG GGGTAT GGGTAT GTCGAT GGCTGA-9604), allowing to hold together the double DNA chain during the following temperature gradient electrophoresis (TGGE) (Rojas-Oropeza et al. 2012).
For TGGE analysis, stapled amplifications of each gene were applied on acrylamide gels. A bisacrylamide (37.5:1) gradient from 56 to 66 ºC with 6 % acrylamide was used for 13 h at 60 V. After the electrophoresis, gels were revealed in a bath of SYBR-Gold with a concentration of 1:1000 (v/v) for 1 h. Digital images were analyzed in the Quantity One software from Bio-Rad Laboratories.
Sequencing and computer analysis
For DNA sequencing, visible bands were excised, resuspended in 20 µl of distilled water, and incubated for 24 h at 4 ºC. Selected DNA-bands were purified with Qiaex II Gel Extraction Kit (Quiagen). The nucleotide sequences of the DNA-band were determined by automated DNA sequencing by using the dideoxy chain-termination method and the ABI model 373A sequencer stretch (Applied Biosystems, Perkin-Elmer) of the Instituto de Fisiología Celular, UNAM. Each sequence was compared with sequences available in databases (GenBank, Blast, NCBI). A selection of Anammox bacteria from soil, wastewater treatment and marine sediment was included in the tree analysis (Nacional Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/). Derived nucleotide sequences of hzoA were aligned with nucleotide sequences of equivalent length using the ClustalW Multiple alignment software of Mega 7.0.21. The tree analysis was performed with Mega 7.0.21 (Kumar et al. 2016). The tree was reconstructed using the neighbor-joining method by Saitou and Nei (1987) and tree topology was evaluated by bootstrap analysis using 100 replicates.
Anammox kinetics
Forty grams of soil were placed in 125 mL glass flasks and fed with an Anammox metabolism specific medium: KH2PO4 3.0 g/L, K2HPO4 4.5 g/L, NH4Cl 2.96 g/L, NaNO2 1.5 g/L and 1.5 mL/L of culture medium of trace elements (EDTA 5 g/L, CaCl2∙2H2O 5.54 g/L, CuSO4∙5H2O 1.57 g/L, (NH4)6Mo7O24∙4H2O 1.1 g/L, CoCl2∙6H2O 1.6 g/L, FeCl3 5.0 g/L, MnCl2 5.0 g/L, MgCl2 5.0 g/L). The different soils were then complemented with distilled water up to saturation (anaerobic conditions) according to the measured water-holding capacity (Table I). Finally, each bottle was hermetically sealed with rubber top and aluminum ring and air was purged with helium gas for 5 min. The kinetics occurred then in an incubator at 23 ºC (average temperature of the studied sites) for a total period of 229 days.
During the whole experiment, a total of three 16-day kinetics were carried out (kinetic 1, 2 and 3). Between each kinetic, microcosms were regularly fed with 2 mL of an Anammox metabolism specific culture medium supplemented with a carbon source: KH2PO4 3.0 g/L, K2HPO4 4.5 g/L, NH4Cl 0.82 g/L, NH4HCO3 0.085 g/L, NaNO2 1.5 g/L and 1.5 mL/L of culture medium of trace elements (EDTA 5 g/L, CaCl2 ∙ 2H2O 5.54 g/L, CuSO4 ∙ 5H2O 1.57 g/L, (NH4)6Mo7O24 ∙ 4H2O 1.1 g/L, CoCl2 ∙ 6H2O 1.6 g/L, FeCl3 5.0 g/L, MnCl2 5.0 g/L, MgCl2 5.0 g/L).
At each point of the kinetics, 20 mL of gas were extracted from the soil flasks and injected in 25 mL helium purged glass bottles for gas analysis. Nitrates, nitrites and ammonium were extracted from 10 g soil using a 0.5 M NaSO4 solution.
Quantification of different nitrogen species
Measurement of concentrations of dinitrogen in gas samples was performed using a GOW-MAC gas chromatograph equipped with a thermal conductivity detector.
Nitrate, nitrite and ammonium concentrations were determined with a Skalar segmented flow analyzer based on the cadmium reduction method. Determinations of nitrite concentration alone were done through a diazo-coupling reaction producing a red-purple coloration absorbing light at 540 nm. Determinations of ammonium concentration were based on the modified Berthelot reaction. Quantification of each nitrogen species was characterized by triplicate.
RESULTS AND DISCUSSION
Presence of the Anammox functional gene hzoA in alkaline saline-sodic soils of the former Lake Texcoco
Following specific amplification of the studied gene, two hzoA partial gene sequences were obtained in the salinity 1. These sequences were individually analyzed using BLAST, to obtain sequences with the highest levels of similarity and allow construction of the dendrogram (Fig 1).
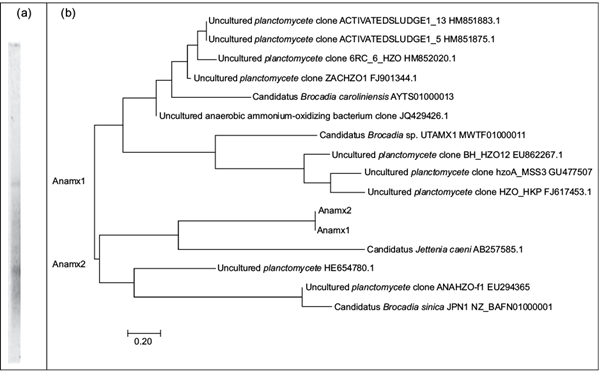
Fig 1 (a) TGGE analysis of the gene amplified with the hzoA specific primers, and (b) dendrogram of the hzoA gene fragments, both present in soils sampled at the former Lake Texcoco. Comparison of temperature gradient gel electrophoresis sequences and sequences from GenBank. Phylogenetic distances were determined analyzing neighbor-joining.
The diversity of the hzoA gene is low, as it presents a range-weighted richness (R r ) of 0.12:
where N is the total number of bands and Tg the gradient of temperature (Marzorati et al. 2008). The fragments Anamx1 and Anamx2 are phylogenetically close to the bacterium Candidatus Jettenia caeni of the phylum Planctomycetes, from an anaerobic ammonium- oxidizing enrichment culture (AB257585.1), followed by an uncultured Planctomycetes from the Baltic Sea (HE654780.1) (Shimamura et al. 2007, Klawonn et al. 2012). Irrigation with wastewater from Mexico City, for more than a century, could explain the exogenous origin of these operational taxonomic units (OTU). However, considering that the phylum Planctomycetes was aggregated with a very wide range of natural environments (Klawonn et al. 2012, Pajares and Bohannan 2016, Wiegand et al. 2018), it is not possible to determine whether these two OTU represent indigenous or exogenous microorganism phylotypes.
The hzoA gene is observed only in the lower salinity (salinity 1). Wang et al. (2020) observed that the dominance of bacterial groups is related to salinity: an increase in salinity changes the dominance of Planctomycetes, Proteobacteria and Bacteroidetes to Proteobacteria and Firmicutes.
Activity of the Anammox function in the alkaline saline-sodic soil of former Lake Texcoco
The Anammox kinetics was performed in three steps: the first step at the initial time, the second step after 40 days of enrichment and the third after 229 days of enrichment. The presence of Anammox microorganisms presents a low diversity. An experiment of enrichment culture was initiated in microcosms. Of the three 16-day kinetics monitored during the 229 days enrichment experiment, only the third one allowed to observe significant consumption of ammonium and nitrite, with a significant production of dinitrogen. It is possible to compare the consumption and production rates of the different nitrogen species during the exponential phase of the third kinetics, with the average rates of the three kinetics (Table II).
TABLE II AVERAGE CONSUMPTION AND PRODUCTION RATES OF THE DIFFERENT NITROGEN SPECIES DURING THE THREE KINETICS AND DURING THE EXPONENTIAL PHASE OF THE THIRD KINETICS.
| Salinity 1 | Salinity 2 | |||||
| Site 1 | Site 2 | Site 3 | Site 1 | Site 2 | Site 3 | |
| Average of kinetics 1 and 2 exponential phase slope (mg/kg dry soil/d) | ||||||
| NH4 + consumption | 1.98 | 2.02 | 2.13 | 3.32 | 3.04 | 1.92 |
| NO2 - consumption | 1.26 | 1.35 | 1.19 | 1.09 | 1.21 | 1.23 |
| N2 production | 6.39 | 3.82 | 5.54 | 2.29 | 2.71 | 2.85 |
| Kinetics 3 exponential phase slope (mg/kg dry soil/d) | ||||||
| NH4 + consumption | 16.70 | 15.07 | 13.37 | 64.81 | 37.93 | 15.27 |
| NO2 - consumption | 1.99 | 2.39 | 1.35 | 1.13 | 3.24 | 4.99 |
| N2 production | 57.55 | 98.80 | 27.17 | 24.72 | 36.27 | 32.09 |
| p-values (N2 production between kinetics 1-2 and 3) | 0.00001 | 0.04033 | 0.00759 | 0.00267 | 0.00189 | 0.00025 |
The production of N2 gas and the concentrations of ammonium (NH4 +) and nitrite (NO2 -) in microcosm soils of the studied salinities are presented in figures 2 and 3. The production of N2 gas is observed in microcosm (Fig 2). Kinetics carried out in sterilized soils (not shown) do not follow this trend, implying that this gas production is indeed induced by anaerobic microbial activity. It can also be noticed that the production of gas in the sites of salinity 2 is at least as significant as in the soil of salinity 1.
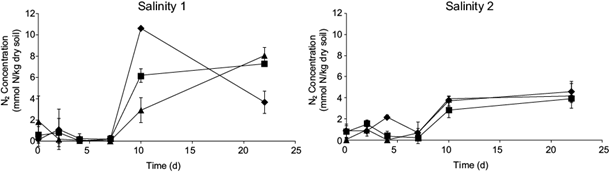
Fig 2 N2 production in a microcosm environment during the third Anammox kinetics for the three sites with each salinity (after 229 days of Anammox enrichment): ■ site 1; ♦ site 2; ▲ site 3. Salinity 1, low conductivity soils; salinity 2, high conductivity soils. Determination by triplicate. ANOVA with p < 0.05.
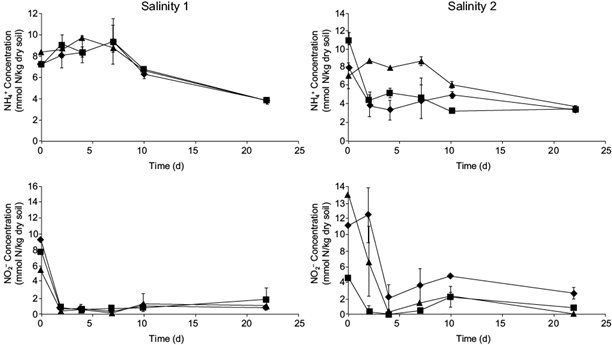
Fig 3 NH4 + and NO2 - concentrations in a microcosm environment during the third Anammox kinetics for the three sites with each salinity (after 229 days of Anammox enrichment): ■ site 1; ♦ site 2; ▲ site 3. Salinity 1, low conductivity soils; salinity 2, high conductivity soils. Determination by triplicate. ANOVA with p < 0.05.
As expected in the case of Anammox activity, NH4 + and NO2 - concentrations followed the opposite trend, a nitrogen removal. While, in each case, NO2 - practically disappeared completely in less than two days, NH4 + showed an average decrease of 54 % during the kinetics, both in the case of each salinity soils (Fig 3). After the Anammox enrichment period of 229 days, the stoichiometric conversion of NO2 - and NH4 + to N2 gas is complete in the studied soils, based on the Anammox process where 1 mole of NH4 + resulted in 1.02 moles of N2 (Saricheewin et al. 2010). These measurements highlight the association of NO2 - and NH4 + consumption with the production of N2 gas. Each time, the limiting element is NO2 -, as the whole nutrient is practically consumed since the fourth day of kinetics for each site of both salinities. It might be possible to consider that Anammox speed is slightly higher in the case of salinity 1 soils.
Therefore, in this study, the presence of a functional gene hzoA confirmed the Anammox process in saline-sodic extremophile soil of the former Lake Texcoco. Our hypothesis was also confirmed by stoichiometric analysis: Anammox activity was present in the two salinities, which is related to the presence of Anammox microorganisms. We are facing a test that strengthens the theory of the influence of the physical and chemical conditions of a saline-sodic soil on the assembly and co-occurrence of the microbial community (Guan et al. 2021). The presence and diversity of microorganisms can occur in the same process, in different times under similar conditions: hydraulic pulse, anaerobiosis, redox potential and nutrient availability.
CONCLUSIONS
The sodium saline soil of the former Lake Texcoco contains the Anammox gene with low diversity, phylogenetically close to the phylum Planctomycetes. Furthermore, Anammox activity was observed after an enrichment period with a specific culture medium for the Anammox process. The existence of microorganisms with Anammox activity in a saline-sodic alkaline soil is of great interest to understand the nitrogen cycle in extremophile soils.











 text new page (beta)
text new page (beta)


