INTRODUCTION
Both Klebsiella pneumoniae and Serratia marcescens are opportunistic pathogens causing nosocomial infections (NI), such as pneumonia, meningitis, bacteremia, endocarditis, conjunctivitis, and urinary tract and wound infections.1,2 Infections caused by S. marcescens or K. pneumoniae have been linked with various hospital sources, such as medical devices, milk, topical and intravenous solutions, liquid soap and air-conditioning systems.1-3 Both bacterial species often cause nosocomial outbreaks and are also commonly resistant to cephalosporins (through. the production of extended-spectrum β-lactamases, ESBLs) and aminoglycosides2-4 ESBLs have a serine moiety in their active sites, and can hydrolyze the β-lactam ring of third-generation cephalosporins and monobactams; genes encoding ESBLs can be chromosomal or plasmid-borne.5-7 Since their first characterization, in the 1980's and 1990's in Germany and France, ESBLs have spread worldwide.8 Changes in the amino acid sequence produce novel enzymes with modified, increased spectrum.5,8 The list of ESBL varieties and families is constantly growing; so far, there are 217 enzymes belonging to the TEM family, 184 to the sulfhydryl variable (SHV) family and 18 to the OXA family, for instance (www.lahey.org/Studies).
In Mexico, previous reports have identified SHV-2, -5 and -12, TLA-1 and CTX-M-15 in K. pneumoniae;9-17 and SHV-2, -5 and -12 in S. marcescens.10,15,18
OBJECTIVE
The objective of this study was to describe and characterize the ESBL and associate their presence with the mechanism of the resistance to third generation cephalosporins, which are one of the families of antibiotics used in the treatment of infections by K. pneumoniae and S. marcescens at Instituto Nacional de Pediatría (National Institute of Pediatrics).
MATERIAL AND METHODS
Bacterial isolates. One-hundred and one isolates from blood cultures of different hospitalized patients were analyzed from January 2002 to December 2005; 94 were K. pneumoniae (26 from the surgery ward, 17 from the oncology ward, 14 from the intensive care unit, and 37 from seven other wards) and 7 were S. marces-cens (all from the oncology ward, 2-14 January 2003). Biochemical identification was performed using the API 20E (bioMérieux) system.
Antibiotic susceptibility assays. Susceptibility towards ampicillin (AMP), amikacin (AMK), aztreonam (ATM), ceftazidime (CAZ), ciprofloxa-cine (CIP), cefotaxime (CTX), cefuroxime (CXM), cefepime (FEP), cefoxitin (FOX), gatifloxacin (GAT), gentamicin (GEN), imipenem (IPM), le-vofloxacin (LVX), tobramycin (TOB), ofloxacin (OFX), streptomycin (STR), trimethoprim-sulfamethoxazole (SXT), tetracycline (TET), ticarcillin (TIM), and piperacillin-tazobactam (TZP) was assessed by the disk diffusion method, following Clinical Laboratory Standard Institute (CLSI) guidelines19 and using Becton-Dickinson disks. Minimum inhibitory concentration (MIC) was inferred from the diameter of inhibitory halos using the BIOMIC System (Giles Scientific).
ESBLs phenotypic detection. The double-disk method,19 using CTX and CAZ disks, alone and in combination with a clavulanic acid disk was used to detect ESBLs. Escherichia coli ATTCC25922 was used as negative control and as positive control K. pneumoniae ATCC700603, following the CLSI standards. The production of inducible AmpC β-lactamases was assessed by a double-disk method20 using CAZ and FOX disks.
Genotyping by pulsed field gel electrophoresis (PFGE). Chromosomal DNA, digested with AbaI and SpeI (Gibco), was resolved in a 1% agarose gel using a CHEF Mapper XA System (Bio-Rad) running for 17 h at 6 V/cm, with pulses of 2-17 s, as previously reported.21 Gels were stained with ethidium bromide and visualized on a Gel-Doc system (Bio-Rad). Images were interpreted according to22Isoelectric focusing (IEF) of β-lactamases. The isoelectric point (pI) of identified ESBLs was assessed as described by Mathew et al.23 Briefly, extracts obtained by sonication24 were run in ampholine-containing gels (Pharmacia Phast) with a 3-9 pH gradient, and run in a PhastSystem system (Pharmacia). β-lactamase activity in the IEF gel was determined by using nitrocefin (500 mg/mL) in combination with CAZ (8 mg/mL), covering the gel agar containing E. coliJ53-2 (CAZ-sensitive) 106 CFU/mL and incubating at 37°C for 16-24 h.
Plasmid extraction. Plasmids were extracted by alkaline lysis25 and visualized by electrophoresis on 0.7% agarose gels stained with ethidium bromide and checked under UV light.
Conjugal transfer. Using CAZ-resistant K. pneu-moniae and S. marcescens clinical isolates as donors, and E. coli J53-2 (rifampicin resistant) as recipient, conjugation experiments were performed according to Miller.26 Transconjugant bacteria were selected on LB agar supplemented with rifampicin (200 pg/mL) and CAZ (1 pg/mL), identified with API-20E, and CAZ-resistance phenotype confirmed by the double-disk method, as described above.
Detection of blaSHV by PCR. Plasmid DNA extracted as described above, was used as template for PCR reactions using primers 5'-GGGTAATTCTTATTTGTCGC-3 ' and 5'-TTAGCGTTGCCAGTGCTC B-3' for the blaSHV genes. Reaction conditions were: one cycle of 95°C/3 min; 30 cycles of 95°C/30 sec, 55°C/30 sec, 72°C/30 sec; and one cycle of 72°C/7 min.27 PCR products were resolved by electrophoresis in 1% agarose gels. Amplicons were extracted from gels using the QIAquick Gel Extraction Kit (Qiagen) and sequenced with an automatic ABI Prism 310 DNA sequencer (Applied Biosystems). Sequence analysis was performed using Clone Manager Suite7, DNA-MAN and Clustal W programs.
RESULTS
Resistance prevalence. Resistance of all isolates to different antibiotics is listed in Table 1. Resistance to CAZ was found in 51/94 K. pneumoniae isolates, and in 5/7 S. marcescens isolates.
Table 1 Profile of susceptibility of nosocomial isolates of K. pneumoniae y S. marcescens
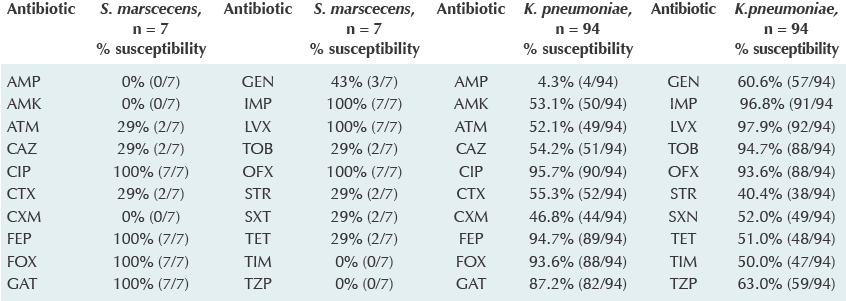
Disk diffusion method, following Clinical Laboratory Standard Institute (CLSI) guidelines.13 20 antibiotics were tested: Ampicillin (AMP), Amikacin (AMK), Aztreonam (ATM), Ceftazidime (CAZ), Ciprofloxacine (CIP), Cefotaxime (CTX), Cefuroxime (CXM), Cefepime (FEP), Cefoxitin (FOX), Gatifloxacin (GAT), Gentamicin (GEN), Imipenem (IPM), Levofloxacin (LVX), Tobramycin (TOB), Ofloxacin (OFX), Streptomycin (STR), Trimethoprim-sulfametoxazole (SXT), Tetracycline (TET), Ticarcillin (TIM), Piperacillin (TZP).
ESBL-production phenotype. All 48 CAZ-resistant isolates tested positive for ESBL production, using the double-disk test, and none for AmpC β-lactamase.
PFGE genotyping. While all 43 CAZ-resistant K. pneumoniae isolates had a distinct PFGE pattern, all 7 CAZ-resistant S. marcescens had an identical pattern, suggesting an outbreak (not shown).
Plasmids. CAZ-resistant K. pneumoniae isolates carried one to eight plasmids, whereas S. marcescens isolates had two to five. None of the isolates had the same plasmid profile, but all of K. pneumoniae share a plasmid of approximately 48 kb.
Conjugal transfer. CAZ-resistant K. pneumoniae isolates successfully transferred the ~48-kb plasmid to a recipient E. coli J53-2, along with the CAZ-resistance phenotype. Experiments using S. marcescens isolates did not yield CAZ-resistant transconjugants.
Isoelectric focusing. CAZ-resistant K. pneumoniae and S. marcescens isolates, and E. coli transconjugants, had two enzymes, with pI of 5.4 and 8.2. The CAZ-hydrolyzing activity was detected in the pI 8.2 enzyme in all strains tested. This pI corresponds to the SHV family.28,29
blaSHV detection by PCR and sequencing. The amplification products using primers for blaSHV genes, in CAZ-resistant clinical isolates and trans-conjugants, showed the expected 900 bp size. Selected amplicons were sequenced; sequence alignments with gene blaSHV¡ (access number AF148850.127 30 ) and blaSHV5 (access number X55640.1.28. 31 ) A 100% homology with SHV-5 type ESBL was found in all analyzed sequences.
DISCUSSION
The prevalence of ESBL-producing K. pneumoniae varies widely: 7.5% in North America, 13.3% in Europe, 22.4% in the Asia-Pacific region, 44% in Latin America, 48.5% in Turkey, 51% in China and 72% in India.1,4 ESBL-producing K. pneumoniae isolates reported here amount to 46%. The prevalence of ESBL-producing organisms is supposed to be related to the inappropriate use of antibiotics, and to the policies on antibiotic usage in each region;32 In México, antibiotics are among the most consumed drugs leading the Latin American region in antibiotic usage up to year 2005.33
The similarity in PFGE profiles among all CAZ-resistant S. marcescens isolates, strongly suggests an outbreak in the oncology ward; this kind of outbreaks are often caused by cross-contamination by medical staff,4 and have a mortality rate of 10-20%.2,4,34 Although the number of isolates reported here is small, two of these seven isolates were related to fatal outcomes, hence a 29% mortality rate. On the other hand, CAZ-resistant K. pneumoniae isolates all had distinct PFGE profiles, but they all carry a similar-size, conjugative plasmid bearing the bla gene, suggesting that the plasmid has been transferred between different K. pneumoniae strains within the hospital in a manner consistent with the notion of epidemic plasmids, outlined nearly 30 years ago.35 Such mobile elements can easily spread antibiotic resistance genes among a variety of bacterial species and across wide geographical areas. As the same blaSHV5 gene was found in all CAZ-resistant S. marcescens isolates, it is even possible that the conjugative plasmid mentioned above was also transferred to a S. marcescens strain within the hospital, starting then an outbreak. Should this be the case, stringent monitoring of the emergence of resistance plasmids in K. pneumoniae in hospital environments could prevent the occurrence of outbreaks caused not only by that bacterial species, but also by other enteric bacteria acting as conjugal recipients. In order to optimize the use of molecular epidemiological monitoring in hospitals from low-income countries, it is important to assess which bacterial species are more likely to act as resistance and transfer "hot spots", so that early detection of dangerous phenotypes could prevent hospital-wide colonization and outbreaks.
CONCLUSION
We found SHV-5 ESBL to be the most common underlying cause of third-generation cephalosporin resistance among K. pneumoniae and S. marcescens isolates causing sepsis in hospitalized children at our institution. The SHV-5 ESBL gene appears to reside in a highly mobile plasmid, capable of spreading among different K. pneumoniae clones and perhaps even to S. marcescens. We suggest designing control programs antibiotics limit the spread of multidrug-resistant bacteria and the spread of conjugative plasmids among strains.











 text new page (beta)
text new page (beta)


