Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.37 n.3 Texcoco Sep. 2019 Epub Sep 30, 2020
https://doi.org/10.18781/r.mex.fit.1906-6
Scientific articles
Identification and cloning of three endogenous genes that may confer resistance to pathogens in citrus including CLas and CTV
1 RGP-Fruticultura, Fitopatología, RGBP-Semillas, Colegio de Postgraduados, Km 36.5 Carretera México-Texcoco, Montecillo, Estado de México, CP 56230;
2 Horticultural Sciences Department, University of Florida, Gainesville, Fl, 32611, USA;
Candidatus Liberibacter asiaticus (CLas), causal agent of Huanglongbing disease (HLB), and the Citrus tristeza virus (CTV)constitute a serious threat to Mexican citriculture because its endemic status in limes in several regions of the Pacific coast and also for the recent detection of severe strains, respectively. The identification and cloning of some genes related to the Systemic Acquired Resistance (SAR) of the plant could help to confront the diseases as an integrated management strategy. This work had the objective of cloning specific genes inherent to the plant as a first step to obtain transgenic citrus plants putatively resistant to CLas. The genes were selected based on the response of citrus plants to a wide range of pathogens, they were; a regulating gene for salicylic acid signaling (SA) as the inducer gen Azelaic Acid Induced 1 (AZI-1), a Citrus Non-Race-Specific Disease Resistance 1 (CsNDR-1) and a pathogenesis related protein 1 gen (PR-1). These three genes (AZI-1, PR-1 and CsNDR-1) were independently inserted into pUC118-FMV-Poly-2-1, subsequently they were subcloned individually into the vector pCAMBIA 2301 while gene AZI-1 was only cloned into pCAMBIA 2201. The CsNDR-1 construct was cloned into Agrobacterium tumefaciens strains EHA5 and AgL1 while genes AZI-1 and PR-1 could not be inserted into these strains of Agrobacterium. These constructions are the first step to be able to generate transgenic citrus as an alternative to face the HLB.
Keywords: SAR; plasmids; AZI-1; PR-1 and CsNDR-1
Candidatus Liberibacter asiaticus (CLas), agente causal del huanglongbing (HLB), y el Citrus tristeza virus (CTV) constituyen amenazas para la citricultura mexicana por su condición endémica en cítricos agrios de algunas regiones del pacífico y de focos recientes de razas severas, respectivamente. La identificación y clonación de genes relacionados con la Resistencia Sistémica Adquirida (RSA) de la planta podrían coadyuvar como parte de una estrategia de manejo integrado. Este trabajo tuvo como objetivo desarrollar construcciones con genes específicos inherentes a la planta como primera etapa en la generación de plantas transgénicas resistentes a CLas. Los genes seleccionados con base a la respuesta de cítricos a una amplia gama de patógenos, fueron AZI-1, 1-inductor del ácido ‘Azelaic’ y CsNDR-1, 1-resistencia no específica a enfermedades en cítricos, ambos genes reguladores de la señalización del ácido salicílico (SA), y el gen asociado con la síntesis de proteína 1 relacionada con la patogenicidad (PR-1). Los tres genes (AZI-1, PR-1 y CsNDR-1) se insertaron independientemente en pUC118-FMV-Poly-2-1. Posteriormente estos tres genes fueron subclonados individualmente en el vector pCAMBIA 2301, mientras que en pCAMBIA 2201 se clonó el gen AZI-1. La clonación de los genes en los vectores pCAMBIA2301 y pCAMBIA2201 permitió incorporar a las construcciones un gen de selección de transformación (35S-nptII-35S) y un gen reportero (35S-GUS-NOS). La construcción CsNDR-1 fue clonada en las cepas EHA5 y AgL1 de Agrobacterium tumefaciens. Los genes AZI-1 y PR-1 no se lograron integrar a estas cepas de Agrobacterium. Estas construcciones representan el primer paso para generar cítricos transgénicos como una alternativa para enfrentar a CLas y CTV.
Palabras clave: RSA; plásmidos; AZI-1; PR-1 y CsNDR-1
The CTV is a subendemic pathogen in Mexico and a recent outbreak of a severe race has been detected in some municipalities of the Gulf of Mexico. The Citrus tristeza virus is an organism regulated by the official division since the 1980s but has remained symptomless. Conversely, Huanglongbing (HLB) or yellow dragon, also known as citrus greening (enverdecimiento de los cítricos), was detected in 2009 and spread rapidly in Mexico. Huanglongbing is a disease caused by a Gram-negative Candidatus liberibacter bacterium (Gottwald, 2007). Three species are known: Candidatus Liberibacter asiaticus (CLas), Candidatus Liberibacter africanus (CLaf) and Candidatus Liberibacter americanus (CLam); Candidatus Liberibacter asiaticus (CLas) is the only one reported in Mexico. CLas attacks all citrus, affects the productive physiology of young and adult plants, including hybrids, by producing moderate-to-severe symptoms and is distributed in almost all the citrus producing countries (Bové, 2012; Hall et al., 2012). This systemic bacterium multiplies and is transported by the phloem and requires insect vectors to spread the disease, the most important being Diaphorina citri (Hall et al., 2012; Ichinose et al., 2010).
Mora-Aguilera et al. (2014) reported a variable level of epidemic intensity with 26% incidence in Brazil and up to 100% in China. The same authors reported it could cause production losses of 42% in sweet orange, 62% in Mexican lemon and 17.3% in Persian lime under the citrus production conditions in Mexico. Due to the epidemic character of the disease, since 2008 the Mexican government has supported a national program for controlling the disease based on eradicating infected plants, controlling the vector and using certified materials, which has made it possible to control the disease impact and identify endemic and subendemic regions such as Colima, Michoacán and Nayarit. Since no resistant varieties are currently available, measures for controlling the vector and an effective agronomic management with special attention on nutrition and irrigation have made it possible to maintain profitability in commercial plantations (Mora-Aguilera, G. 2018. Data not published; Fu et al., 2016). At the experimental level, there are several physical and chemical alternatives for controlling CLas (Zhang et al., 2014; Vazquez-Garcia et al., 2013; Zhang et al., 2012; Hoffman et al., 2012; Ding et al., 2008). Some of these methods have not produced consistent results, and others, including the use of antibiotics, are not environmentally or economically viable. The development of transgenic citrus varieties as an alternative for rapidly developing resistance has proven to be viable against CTV, but the use of genes exogenous to the plant has limited its commercial implementation. One alternative is to use CRISPR, a gene editing technique that is still being developed, and endogenous genes associated with plant defense mechanisms.
The activation of these mechanisms starts by recognizing effectors such as flagelin, chitins, glucloproteins and lipopolysaccharides known as pathogen-associated molecular patterns (PAMPs). The proteins that recognize these effectors are known as resistance proteins (R). This specific response mechanism is called gene-for-gene resistance sensu Flor (Gao et al., 2015; Nishimura and Dangl 2010; Belkhadir et al., 2004). The type of resistance conferred by R proteins only in the presence of a pathogen, is called systemic acquired resistance (SAR) (Gómez and Mejía, 2011).
Recently, analyses of citrus transcriptome in response to CLas, CLam and CTV infection have demonstrated that several genes are over-expressed in response to infection, while others are restrained (Hu et al., 2017; Rawat et al., 2017; Da Graca et al.,2016; Fu et al., 2016). The multiple genes consistently associated with RSA include CsNDR-1, AZI-1 and PR-1, which are expressed in response to phytopathogens of citrus and other crops (Hu et al., 2017; Da Graca et al., 2016; Lu et al., 2013; Pajerowska et al., 2013: Yu et al., 2013; Van Loon and Van Strien, 1999). Genes activated by the presence of phytopathogens have been identified, which offers the possibility of ensuring the continuous and regular expression of those genes in the plant through genetic engineering in order to avoid infection processes. Therefore, the objective of this study was to identify and isolate CsNDR-1, AZI-1 and PR-1 using genetic engineering, clone them in transformation vectors and introduce them into two Agrobacterium tumefaciens strains for future implementation in citrus genetic transformation using endogenous genes in order to induce resistance to CLas and CTV.
Materials and methods
Biological material. Plants of Arabidopsis thaliana and sweet orange (Citrus sinensis) were grown in a mixture of peat moss-perlite substrate (60:40) under greenhouse conditions at 27-30 °C temperature and 2243 masl, in Montecillo, Texcoco, State of Mexico.
Bioinformatic analysis. The consensus sequences in the Arabidopsis (Pajerowska M.K. et al., 2013; Yu et al., 2013) and C. sinensis database (Lu et al., 2013) were found using the BLAST (Basic Local Alignment Search Tools) program, and the sequence with the highest similarity (Query) was selected. The analysis of alignment of the conserved sequences of the reported genes was conducted using the Mega6 program. The designed primers were analyzed using the Oligo Analyzer Tool and Primer Questo Tool (IDT, Integrated DNA Technologies) programs, and NEBcutter (BioLabs) was used to detect the possible formation of dimers and identify the restriction sites in the primers.
DNA extraction. To extract DNA from the AZI-1 and PR-1 gene isolates, 0.1 gram of mature A. thaliana leaves was used following the method of 2% CTAB (Doyle and Doyle, 1987). To extract DNA from the CsNDR-1 isolate, young leaves of C. sinensis and the QIAGEN DNAeasy kit were used following the manufacturer’s instructions. DNA concentration was measured using 1 µL of the sample in a Nanodrop (Thermo SCIENTIFIC, NANODROP 2000).
Design of primers. The gene search was conducted using BLAST. For the CsNDR-1 gene the following sequence was found: gi|568842565|ref|XM_006475151.1| PREDICTED: Citrus sinensis protein CsNDR-1-like (LOC102630232), mRNA. Based on the sequence found, one ‘Forward’ primer and one ‘Reverse’ primer were designed, both containing a series of 6 A/T followed by the sequence of the restriction enzyme selected for cloning plus the Kozac ACC sequence, and, finally, the primer sequence.
The following record of the AZI-1 gene was found: gi|30682133ǀrefǀNM_117317.4ǀ Arabidopsis thaliana azelaic acid induced 1 (AT4G12470), mRNA, and for PR-1 giǀ 17381133ǀAY064023ǀ Arabidopsis thaliana putative pathogenesis-related PR-1 protein (At2g14610), mRNA. Based on the sequences found, the ‘Forward’ and ‘Reverse’ primers were designed for each gene.
Gene identification and amplification. Gene detection and amplification through end-point PCR was performed using specific primers for each gene, including restriction points for cloning (Table 1).
To design the primers, restriction sites of Apa1 (GGGCCC) and Spe1 (ACTAGT) were incorporated, as well as several adenines following the restriction sites. For this, we used a final volume of 25 µL of a mixture containing 5 µL of DNA, 2.5 µL of 10X PCR Buffer (Invitrogen), 1 µL of MgCl2 50 (Invitrogen), 0.5 µL of dNTPs 10 mM (Promega), 1 µL of each primer (10 pmol), RNAsa/DNAsa-free H2O (Invitrogen) and 0.2 µL of Taq polymerase (Invitrogen). The amplification program consisted of 35 cycles for AZI-1 and PR-1: 95 °C for 30 s, 68 °C for 30 s and 68 °C for 30 s. For CsNDR-1, 35 cycles: 95 °C for 30 s, 66 °C for 30 s and 68 °C for 40 s. The three genes had an initial pre-denaturation stage at 95 °C for 2 min and a final extension stage at 68 °C for 5 min.
The PCR products were analyzed in 1% agarose gel, with 5 µL of the PCR product and 3 µL of load buffer (Promega). The gel was run at 90 volts, 400 amperes for one hour, and was dyed in ethidium bromide for 5 min. The PCR products that amplified at the expected size were purified using the QIAquick® PCR Purification kit and sequenced to corroborate the isolation of the tested gene before using engineering.
Vector construction. The pUC118-FMV-Poly-2-1 vector (Febres et al., 2003) was grown in three millimeters of 2xYT liquid culture medium (yeast extract 10 g L, tryptone 16 g L, NaCl 5 g L) containing 100 µg mL-1 of ampicillin (amp) at 37 °C for 12 h and then the plasmid was isolated using the QIAprep®Spin miniprep kit, following the manufacturer’s instructions. The purified AZI-1, PR-1 and CsNDR-1 genes, as well as the pUC118-FMV-Poly-2-1 isolated vector, were digested with Apa1 and Spe1 restriction enzymes at the temperature and time recommended for each enzyme. The double digestion of the three genes and vector pUC118-FMV-Poly-2-1 produce a single fragment, so they were purified using a QIAquick® PCR Purification kit, and then ligated for 12 h at 4 °C. The digestion products were cloned in Escherichia coli DH5α (Invitrogen) using 80 to 100 ng µL-1 of DNA by electroporation under one pulse conditions at 1.8 kV (Bio-Rad, 165-2100). The bacteria that were supposedly transformed were sown in a 2xYT solid culture medium containing 100 µg mL-1 of amp; three aliquots of transformed cells (10, 20 and 50 µL) were tested using a sterile microinjector to distribute them on the medium surface. After 12 h at 37 °C, several individual colonies formed and were analyzed using PCR with the previously mentioned primers in order to corroborate the presence of the genes. The PCR products were run in 0.8% agarose gel at 100 volts for 30 min.
Table 1 Primers designed to isolate and clone the AZI-1, PR-1 and CsNDR-1 genes.
| Cebadorx | Secuenciay | Ampliconz |
|---|---|---|
| AZI1 F | 5´ AAA GGGCCCATGGCTTCAAAGAACTCAGCCTCTCTTG-3´ | 486pb |
| AZI1 R | 5´-AAA ACTAGTTCAAGCACATTGGAAACCAGATGGAAGC-3´ | |
| PR-1 F | 5´AAA GGGCCCATGAATTTTACTGGCTATTCTCGATTTTTAA-3´ | 486pb |
| PR-1 R | 5´-AAA ACTAGTTTAGTATGGCTTCTCGTTCACATAATTCC-3´ | |
| CsNDR1 F | 5´-AAA AAA GGGCCCACCATGTCAGAAAACGCCGGTGGGTGCTGC-3´ | 618pb |
| CsNDR1 R | 5´-AAAAAA ACTAGTTTAAGCAAAAATCAAGACAAAAAAATACAAC-3´ | |
x Name of the primers: F:forward, R: reverse.
y Sequence of each primer used.
z Size of the amplicon to obtain each gene.
The positive colonies were sown in three ml of 2xYT liquid culture medium containing 100 µg mL-1 of amp at 37 °C at 230 rpm for 12 h. Using half of the bacterial culture that had grown (1.5 ml), the plasmid was extracted using the QIAprep Spin Miniprep kit from QIAGEN and sent out for sequencing to verify the sequences of the three genes. With the other half of the bacterial cultures, “stocks” were prepared using glycerol and then stored at -80 °C.
To continue inserting the genes of interest now into pCAMBIA transformation vector plasmids, the following procedure was used: the bacteria with the three genes independently contained in pUC118-FMV-Poly-2-1 were grown in 2xYT solid culture medium with 100 µg mL-1 of ampicillin, while those containing the pCAMBIA2201 and pCAMBIA 2301 plasmids were grown in solid 2xYT culture medium with 25 µg mL-1 of chloramphenicol (Cap) or 50 µg mL-1 of kanamycin (Kan), respectively, for 12 h at 37 °C. The plasmids were extracted with a QIAprep® Spin Miniprep kit and then digested with BamH1 and SphI restriction enzymes at the temperature and time recommended for each enzyme. The double digestion products of pUC118-FMV-Poly-2-1 were purified using the QIAgen gel purification kit, while the double digestion products of the pCAMBIA vectors were purified using the Qiaprep Miniprep kit. Product ligation was carried out by incubating them at 4 °C for 12 h. The digestion products were cloned in Escherichia coli DH5α (Invitrogen) using 80 to 100 ng µL-1 of DNA by electroporation under one pulse conditions at 1.8 kV (Bio-Rad, 165-2100). The bacteria that were supposedly transformed were sown in 2xYT solid culture medium containing 25 µg mL-1 of Cap for pCAMBIA2201, or 50 µg mL-1 of Kan for pCAMBIA 2301; three aliquots of transformed cells (10, 20 and 50 µL) were tested using a sterile microinjector to distribute them on the medium surface. After 12 h at 37 °C, several individual colonies formed and were analyzed through PCR using the specific primers in order to corroborate the presence of each of the three genes.
Once more, the positive colonies were individually grown in a liquid culture medium containing the corresponding antibiotics for each plasmid at 37 °C at 230 rpm for 12 h. Similarly, one aliquot was sent out for sequencing, and the other aliquot of bacteria was used to prepare “stocks” that were stored at -80 °C.
Transformation of A. tumefaciens. The pCAMBIA plasmids with the constructions were grown in 2xYT solid culture medium containing 25 µg mL-1 of Cap for pCAMBIA2201, or 50 µg mL-1 of Kan for pCAMBIA 2301 at 37 °C for 12 h, and then the plasmids were extracted using the Qiaprep Spin miniprep kit. The competent EHA 105 and AGL 1 strains of A. tumefaciens (Kayim and Koc 2005; Almeida et al., 2003) were selected for transformation through electroporation (Bio-Rad, 165-2100) under one pulse conditions at 2.5 kV (Bio-Rad, 165-2100). The bacterial cells of the EHA 105 strain that were supposedly transformed were grown in solid YEP culture medium (10 g L of bactopeptone, NaCl 5 g L, yeast extract 10 g L) with rifampicin (Rif) (50 µg mL-1) and Kan (50 µg mL-1), while the cells of the AGL 1 strain were grown in solid YEP culture medium with Rif (50 µg mL-1), Kan (50 µg mL-1) and carbenicillin (Carb) (25 µg mL-1). Aliquots of 50, 100 and 200 µL of the two Agrobacterium strains were tested and incubated for two to three days at 27 °C. The individual colonies that grew were analyzed through end-point PCR using the primers and under the conditions previously described for each gene. The colonies that were positive were sent out for sequencing. The colonies whose transformation and construction were verified through sequencing were grown in liquid YEP culture medium using the corresponding antibiotics before being cryopreserved at -80 °C.
Results and discussion
Identifying genes. The concentration of Arabidopsis DNA was 478.2 ng µL-1 with a value of optical density (O.D.) units of 2.04. The concentration of C. sinensis DNA was 48.7, and its OD reading was 1.7. The PCR products run in agarose gel showed bands of the expected size, that is, 486 pb, corresponding to the AZI-1 and PR-1 genes, and for the CsNDR-1 gene one 618 pb band was obtained (Figure 1). Sequencing of the products showed at least 99% identity.
Cloning genes in the pUC118-FMV-Poly-2-1 plasmid. The purified genes were inserted into the pUC118-FMV-poly-2-1 vector (Febres et al., 2013). Individual E. coli colonies that grew were analyzed through PCR to detect the transformed colonies. For the AZI-1 gene, 18 colonies were analyzed using the AZI-1 F/AZI-1 R primers, and three positive colonies were obtained (Figure 2A, lanes 2, 4 and 10). For the PR-1 gene, 18 colonies were also analyzed using the PR-1 F/PR-1 R primers, and six positive colonies were obtained (Figure 2B, lanes 2, 9, 11 to 13, and 16). For the CsNDR-1 gene, the transformation was done twice, and a total of 35 colonies were analyzed using the CsNDR-1 F/CsNDR-1 R primers; two positive colonies were obtained, and one of them appears in lane 12 of Figure 2C. All the colonies that were positive by PCR were sent out for sequencing, and the colony of each of the AZI-1, PR-1 and CsNDR-1 genes that showed the greatest similarity with the sequence reported in the database of the NCBI GenBank was selected to continue cloning in vector pUC118-FMV-poly-2-1. For the PR-1 gene, colony three showed the highest level of similarity (99%); in the case of the AZI-1 gene, colony one showed 99% similarity, and colony 14 showed 99% similarity in the CsNDR-1 gene. Three constructions were built using the best colonies of each gene inserted between the FMV promoter and the 35S termination signal of pUC118-FMV-Poly-2-1 (Figure 3A, B and C).

Figure 1 Amplification of the AZI-1, PR-1 and CsNDR-1 genes. 1.5% agarose gels with 5 μl of PCR product (lanes 1-2 and 4-5) plus 3 μl of load buffer. M; molecular weight marker (1 kb DNA ladder). A). Lane 1: amplification of the Ara bidopsis AZI-1 gene. Lane 2: negative control of the AZI-1 gene. Lane 4: amplification of the Arabidopsis PR-1 gene. Lane 5: negative control of the PR-1 gene. B). Lane 1: CsNDR-1 negative control. Lanes 2 and 3: amplification of the CsNDR-1 gene.
The genes isolated in this study (CsNDR-1, AZI-1, PR-1) have been consistently described as possible candidates for generating resistance to several pathogens (Lu et al., 2013). These genes are part of the 633 genes that are altered after inoculation with CLas, according to Mafra et al. (2013), and are associated with the plant’s systemic acquired resistance (SAR). Several authors have also identified these genes as part of the plant’s response to CLas and CTV attacks (Hu et al., 2017; Rawat et al., 2017; Da Graca et al.,2016; Fu et al., 2016), and, for this reason, they were selected to develop the constructions.

Figure 2 PCR confirmation of DH5α colonies transformed with the AZI-1, PR-1 and CsNDR-1 genes contained in the pUC118-FMV-Poly-2-1 plasmid. Amplified fragments in 0.8% agarose gel. M: molecular weight marker (1 kb DNA ladder). A). Lane 1: Positive control of the AZI-1 gene. Lanes 2, 4 and 10: Amplification of the expected gene. Lane 19: negative control of the AZI-1 gene. B). lane 1: positive control of the PR-1 gene. Lanes 2, 9, 11, 12, 13 and 16: positive colonies from the amplification of the PR-1 gene. Lane 19: negative control of the PR-1 gene. C). Lanes 1-15: CsNDR-1 gene colonies analyzed. Lane 12: positive sample. Lane 16: negative control.
The pUC118-FMV-Poly-2-1 plasmid was selected because of its small size, and because it produces a large number of copies (500-700). In addition, its multiple cloning site facilitates the integration of DNA fragments within this vector (Febres et al., 2013), and it is also compatible for subsequent cloning within the pCAMBIA vectors incorporating the CaMV35S and/or NOS promoters and terminators of pCAMBIA.
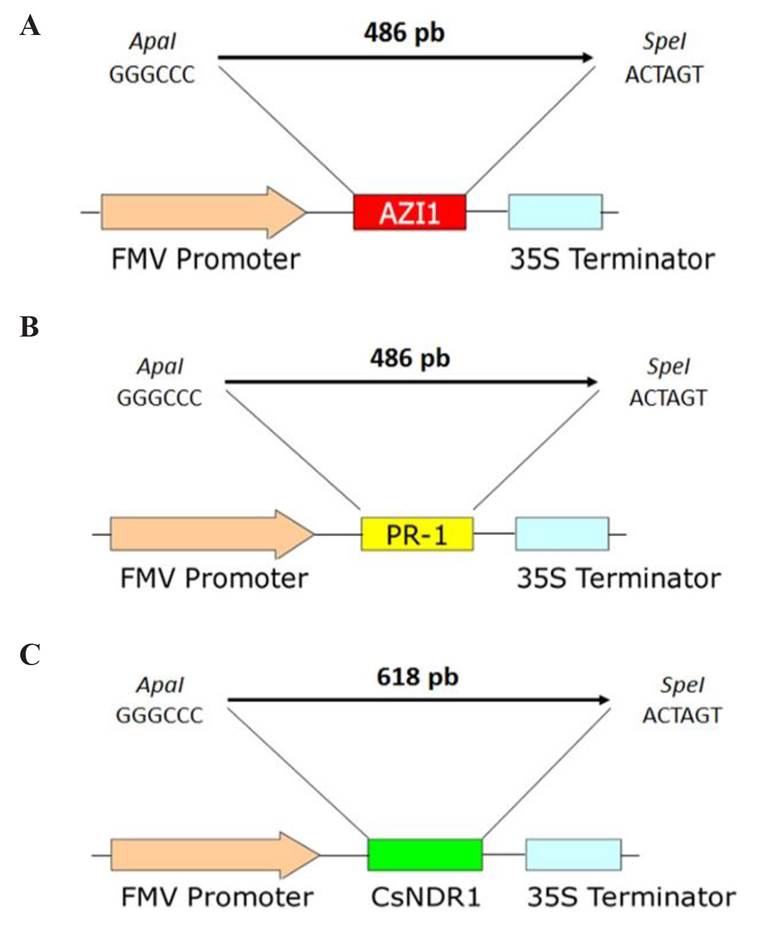
Figure 3 Schematic representation of the constructions of the genes inserted into the pUC118-FMV-Poly-2-1 plasmid. A). AZI-1, B). PR-1, and C). CsNDR-1. The image shows the AZI-1 (red), PR-1 (yellow) and CsNDR-1 (green) genes cloned under the FMV promoter (orange) and their 35S termination signal (blue). The upper part of each construction indicates the size of the amplicon and the restriction enzymes used during cloning.
Cloning in pCAMBIA plasmids. The constructions containing the genes of interest were inserted into vectors pCAMBIA 2201 and 2301. The cloning process was repeated twice, modifying DNA concentrations in the ligation reaction because no transformed colonies with the genes of interest were found. Twenty-one colonies were analyzed in each experiment, for a total of 42 colonies analyzed for each gene. For the AZI-1 gene cloned in pCAMBIA 2201, the concentration of DNA-plasmid was modified in the ligation reaction from 31.8 ng µL-1-108 ng µL-1 to 63.6 ng µL-1-72 ng µL-1; the grown colonies were analyzed by PCR using the AZI-1 F/AZI-1 R primers, and 10 positive colonies were obtained from the second cloning replication (Figure 4A). The AZI-1 gene cloned in pCAMBIA 2301, the DNA concentration in the ligation reaction was from 41.1 ng µL-1-100 ng µL-1 to 82.2 ng µL-1-86.4 ng µL-1; the grown colonies were also analyzed by PCR using the AZI-1 F/AZI-1 R primers, and four positive colonies were obtained from the second experiment (Figure 4B). For the PR-1 gene cloned in the pCAMBIA 2301 vector, the DNA concentration in the ligation reaction was modified from 25.2 ng µL-1-86.4 ng µL-1 to 37.8 ng µL-1-72 ng µL-1, and two hours were added to the incubation step after the electric shock; the grown colonies were analyzed using the PR-1 F/PR-1 R primers, and three positive colonies were obtained in the last experiment (Figure 4C). The constructions amplified a product of 486 pb, as expected. For the CsNDR-1 gene in pCAMBIA, a single test was carried out; 32 colonies were analyzed and only two positive colonies were obtained (Figure 4D).
All the constructions containing the AZI-1, PR-1 and CsNDR-1 genes under the FMV promoter and the 35S terminator were cloned between the neomicin fosfotransferasaII (ntptII) selection gene that confers resistance to kanamycin and the β-glucoronidasa (uidA) reporter gene, known as GUS, of the pCAMBIA 2201 and 2301 plasmids. The nptII transformation selection gene remained with the 35S promoter and terminator, and the GUS reporter gene remained with the 35S promoter and the NOS terminator. The AZI-1, PR-1 and CsNDR-1 genes cloned in pCAMBIA 2301 have the nptII gene as the bacterium selection gene (Figure 5A, C and D). The AZI-1 gene inserted in pCAMBIA 2201 is located under the bacterium selection gene to chloramphenicol (Figure 5 B).

Figure 4 PCR confirmation of DH5α colonies with the AZI-1, PR-1 and CsNDR-1 genes inserted into the pCAMBIA 2201 and 2301 plasmids. Amplified fragments in 0.8% agarose gel. M: molecular weight marker (1 kb DNA ladder). A). AZI-1-pCAMBIA 2201. Lane 1: Positive control of the AZI-1 gene. Lanes 4, 6, 7, 8, 11, 12, 14, 16-21: amplification of the expected gene. Lane 23: negative control. B). AZI-1-pCAMBIA 2301. Lane 1: positive control. Lanes 3, 18, 21 and 22: amplification of the expected gene. Lane 23: negative control. C). PR-1-pCAMBIA 2301. Lane 1: positive control of the PR-1 gene. Lanes 6, 7 and 20: positive colonies from the amplification of the PR-1 gene. Lane 23: negative control. D). CsNDR-1 in pCAMBIA 2301. Lanes 20 and 22: positive colonies for the CsNDR-1 gene. Lane 24: negative control.
The constructions were sent out for sequencing in order to corroborate the identity and orientation of the insertion before being transformed to A. tumefaciens; the ones that were at least 99% homologous were selected. Figure 6 shows 100% identity of colony two in the case of the AZI-1 gene inserted in pCAMBIA2301. The pCAMBIA plasmids are successfully used in the genetic transformation of several plants, including citrus (De Oliveira et al., 2015; Hajeri et al., 2014; Pinheiro et al., 2014).
Transformation of A. tumefaciens. Two A. tumefaciens strains were transformed using the three genes cloned in pCAMBIA and then individual colonies were analyzed through PCR to confirm that they have been transformed. The construction of the PR-1 gene in pCAMBIA2301 in the EHA105 strain developed several individual colonies (Figure 7A), as did the construction of the CsNDR-1 gene in pCAMBIA2301 in the Ag11 strain (Figure 7B). The PCR results confirmed that there were positive colonies for both genes, but the verification through sequencing did not show a high level of identity in the case of the construction with the PR-1 gene. The construction of the CsNDR-1 gene with the FMV promoter and the 35S terminator cloned in pCAMBIA 2301 was successfully inserted into the two A. tumefaciens EHA 105 and AGL 1 strains. Eleven colonies were randomly analyzed through PCR using the CsNDR-1 F/CsNDR-1 R primers, and one positive colony was obtained in the EHA105 strain (Figure 8A), while four positive colonies were obtained for the AGL1 strain (Figure 8B). The low transformation efficiency of Agrobacterium and E. coli is somewhat common in gene construction and cloning experiments. Gelvin (2003) mentioned that many factors may be involved, such as the low competence of the bacterial cells, stressed cells, poor quality DNA, low or high DNA concentration during transformation, the conditions during fragment ligation, the conditions during electroporation, the incubation temperature, and the concentration of antibiotics, among others. The A. tumefaciens EHA 105 strain has been used in citrus genetic transformation and has produced excellent transformation percentages because of its virulence (Hao et al., 2016; Dutt et al., 2015). The AgL1 strain has also shown a high level of transformation in citrus (Shi et al., 2016; Kayim and Koc 2005).

Figure 5 Schematic representation of the construction in pCAMBIA 2201 and 2301. FMV-AZI-1-35S construction inserted into the plasmids A). pCAMBIA 2301, and B). pCAMBIA 2201. C). FMV-PR-1-35S construction, and D). FMV-CsNDR-1-35S construction inserted into the pCAMBIA 2301 plasmid. The genes of interest (red, yellow and green bars) were inserted between the ntptII selection gene that confers resistance to kanamycin with the CaMV (Cauliflower mosaic caulimovirus) promoter and terminator, and the GUS reporter gene under the 35S promoter of CaMV and the nopaline synthase (NOS) terminator.
Conclusions
The AZI-1, PR-1 and CsNDR-1 genes were isolated through PCR using the designed primers using the Arabidopsis thaliana and Citrus sinensis genome that had specific restriction sites for cloning. The isolated and cloned sequences of the AZI-1, PR-1 and CsNDR-1 genes showed 99% similarity compared to the records of the A. thaliana and C. sinensis sequences in the NCBI GenBank database.

Figure 7 A). Transformed colonies of A. tumefaciens EHA 105 with the pCAMBIA 2301 plasmid with the PR-1 gene in YEP culture medium with Rif (50 μl/ml). B). Transformed colonies of A. tumefaciens Agl1 with the pCAMBIA 2301 plasmid with the CsNDR-1 gene in YEP culture medium with Rif (50 μl/ml), Kan (50 μg/ml) and Carb (25 μg/ml). The numbers indicate the colonies that were randomly selected to confirm the presence of the gene using PCR.

Figure 8 PCR of A. tumefaciens colonies. M: Molecular marker (1 kb DNA ladder). A). EHA 105 transformed with the CsN DR-1 gene. Lane 1: positive control. Lane 8: positive colony with the CsNDR-1 gene. Lane 12: negative control. B). AGL 1 transformed with the CsNDR-1 gene. Lane 1: CsNDR-1 positive control. Lanes 2-5: positive colonies with the CsNDR-1 gene. Lane 6: negative control.
The three genes (AZI-1, PR-1 and CsNDR-1) were incorporated between the FMV promoter and the 35S CaMV terminator of the pUC118-FMV-poly 2-1 cloning vector. Cloning the genes in the pCAMBIA2301 and pCAMBIA2201 vectors made it possible to incorporate a transformation selection gene (35S-nptII-35S) and one reporter gene (35S-GUS-NOS) into the constructions. The three genes were then independently cloned into pCAMBIA2301 while gen AZI-1 was cloned only into pCAMBIA2201.
The CsNDR-1 gene was incorporated into the A. tumefaciens EHA 105 and AgL1 strains. Genes AZI-1 and PR-1 could not be integrated into any of the Agrobacterium strains. Identifying, isolating and cloning the CsNDR-1, AZI-1 and PR-1 genes involved in the plants defense mechanisms against a pathogen’s attack may be over-expressed through genetic engineering in citrus and used to induce resistance to several pathogens, including the Candidatus liberibacter bacterium, the causal agent of HLB and Citrus tristeza virus.
Literatura citada
Almeida AB, Mourão-Filho AA, Januzzi-Mendes BM, Pavan A. y Martinelli-Rodriguez AP. 2003. Agrobacterium-mediated transformation of Citrus sinensis and Citrus limonia epicotyl segments. Scientia Agricola 60:1: 23-29. http://dx.doi.org/10.1590/S0103-90162003000100005. [ Links ]
Belkhadir Y, Nimchuk Z, Hubert D, Mackey D and Danglet J. 2004. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. The Plant Cell. 16:10: 2822-2835. https://doi.org/10.1105/tpc.104.024117 [ Links ]
Bové JM. 2012. Huanglongbing and the future of citrus in Sao Paulo, Brazil. Journal of Plant Pathology. 94(3):465-467. [ Links ]
Da Graca JV, Douhan GW, Halbert SE, Keremane ML, Lee RF, Vidalakis G, and Zhao H. 2016. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. Journal of Integrative Plant Biology 58(4):373-387. https://doi.org/10.1111/jipb.12437 [ Links ]
De Oliveira MLP, Stover ED and Thomson JG 2015. The codA gene as a negative selection marker in Citrus. SpringerPlus 4:264: 3-7. https://doi.org/10.1186/s40064-015-1047-y [ Links ]
Ding F, Jin S, Hong N, Zhong Y and Cao Q. 2008. Vitrification-cryopreservation, an efficient method for eliminating Candidatus Liberobacter asiaticus, the citrus Huanglongbing pathogen, from in vitro adult shoot tips. Plant Cell Reports 27(2): 241-250. https://doi.org/10.1007/s00299-007-0467-8 [ Links ]
Doyle JJ and Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of small quantities of fresh leaf tissue. Phytochemical bulletin 19. 11-15. [ Links ]
Dutt M, Barthe G, Irey M and Grosser J. 2015. Transgenic citrus expressing an Arabidopsis NPR-1 gene exhibit enhanced resistance against Huanglongbing (HLB; Citrus Greening). Public Library of Science One. 10:9: 1-17. https://doi.org/10.1371/journal.pone.0137134 [ Links ]
Febres VJ, Niblet CL, Lee RF and Moore GA. 2003. Characterization of grapefruit plants (Citrus paradisi Macf.) transformed with citrus tristeza closterovirus genes. Plant Cell Reports 21: 421-428. https://doi.org/10.1007/s00299-002-0528-y [ Links ]
Fu S, Shao J, Zhou Ch, and Hartung JS. 2016. Transcriptome analysis of sweet orange trees infected with ‘Candidatus Liberibacter asiaticus’ and two strains of Citrus Tristeza Virus. BMC Genomics 17:349 https://doi.org/10.1186/s12864-016-2663-9. [ Links ]
Gao QM, Zhu S, Kachroo P and Kachoroo A. 2015. Signal regulators of systemic acquired resistance. Frontiers in Plant Science 6: 228. https://doi.org/10.3389/fpls.2015.00228 [ Links ]
Gómez S y Mejía Z. 2011. Respuesta de hipersensibilidad, una muerte celular programada para defenderse del ataque por fitopatógenos. Revista Mexicana de Fitopatología. 29:2: 154-164. Disponible en línea: http://www.redalyc.org/pdf/612/61222864007.pdf [ Links ]
Gottwald TR. 2007. Citrus canker and citrus huanglongbing, two exotic bacterial diseases threatening the citrus industries of the western hemisphere. Outlooks on PestManagement 18(6):274-279. https://doi.org/10.1564/18dec09 [ Links ]
Hajeri S, Killiny N, El-Mohtar C, Dawson WO and Gowda S. 2014. Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphorina citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing). Journal of Biotechnology 176:20: 42-49. http://doi.org/10.1016/j.jbiotec.2014.02.010 [ Links ]
Hall DG, Richardson ML, Ammar El-Desouky and Halbert SE. 2012. Asian citrus psyllid, Diaphorina citri, vector of citrus huanglongbing disease. Entomologia Experimentalis et Applicata. 146: 207-223. 10.1111/eea.12025 [ Links ]
Hao G, Stover E and Gupta G. 2016. Overexpression of a modified plant thionin enhances disease resistance to Citrus canker and Huanglongbing (HLB). Frontiers in Plant Science 7: 1078: 1-11. https://doi.org/10.3389/fpls.2016.01078 [ Links ]
Hoffman MT, Doud MS, Williams L, Zhang M and Ding F. 2012. Heat treatment eliminates ‘Candidatus Liberibacter asiaticus’ from infected citrus trees under controlled conditions. Phytopathology. 103:15-22. https://doi.org/10.1094/PHYTO-06-12-0138-r [ Links ]
Hu Y, Zhong X, Liu X, Lou B, Zhou Ch, and Wang X. 2017.Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. Public Library of Science ONE 12(12)e0189229. https://doi.org/a0.1371/journal.pone.0189229. [ Links ]
Ichinose K, Miyazi K, Matsuhira K, Yasuda K, Sadoyama Y, Do HT and Doan VB. 2010. Unreliable pesticide control of the vector psyllid Diaphorina citri (Hemiptera: Psyllidae) for the reduction of microorganism disease transmission. Journal of Environmental Science and Health B. 45:5:466-472. https://doi.org/10.1080/03601231003800263 [ Links ]
Pajerowska MK, Emerine DK and Shahid-Mukhtar M. 2013. Tell me more: roles of NPRs in plant immunity. Trends in Plant Science. 18:7: 402-4011. https://doi.org/10.1016/j.tplants.2013.04.004 [ Links ]
Kayim M and Koc NK. 2005. Improved transformation efficiency in Citrus by plasmolysis treatment. Journal of Plant Biochemistry and Biotechnology 14(1):15-20. https://doi.org/10.1007/BF03263218 [ Links ]
Lu H, Zhang C, Albrecht U, Shimizu R, Wang G and Bowman KD. 2013. Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Frontiers in Plant Science. 4:157:1-10. https://doi.org/10.3389/fpls.2013.00157 [ Links ]
Mafra V, Martins P, Francisco C, Ribeiro-Alves M, Freitas-Astúa J and Machado M. 2013. Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics. 14:247. https://doi.org/10.1186/1471-2164-14-247. [ Links ]
Mora-Aguilera G, Robles GP, López AJL, Flores SJ, Acevedo SG, Domínguez MS, Gutiérrez EA and Loeza KE. 2014. Current situation and perspectives in management of citrus HLB. Mexican Journal of Phytopathology 32(2):108-119. https://doi.org/10.7550/rmb.46314 [ Links ]
Nishimura MT and Dangl JL. 2010. Arabidopsis and the plant immune system. The Plant Journal 61(6): 1053-1066. https://doi.org/10.1111/j.1365-313X.2010.04131.x [ Links ]
Pinheiro TT, Figueira A and Latado RR. 2014. Early-flowering sweet orange mutant “x11” as a model for functional genomic studies of Citrus. BMC Research Notes. 7:511: 1-7. https://doi.org/10.1186/1756-0500-7-511. [ Links ]
Rawat N, Kumar B, Albrecht U, Du D, Huang M, Yu Q, Zhang Y, Duan YP, Bowman KD, Gmitter FGJr, and Deng Z. 2017. Genome resequencing and transcriptome profiling reveal structural diversity and expression patterns of constitutive disease resistance genes in Huanglongbing-tolerant Poncirus trifoliate and its hybrids. Horticulture Research 4,17064. https://doi.org/10.1038/hortres.2017.64 [ Links ]
Shi Q, Febres VJ, Jones JB and Moore GA. 2016. A survey of FLS2 genes from multiple citrus species identifies candidates for enhancing disease resistance to Xanthomonas citri ssp. citri. Horticultural Research 3:16022: 1-11. https://doi.org/10.1038/hortres.2016.22. [ Links ]
Van-Loon LC and Van Strien EA. 1999. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological and Molecular Plant Pathology. 55. 85-97. https://doi.org/10.1006/pmpp.1999.0213 [ Links ]
Vazquez-García M, Velázquez-Monreal J, Medina-Urrutia VM, Cruz-Vargas CD, Sandoval-Salazar M, Virgen-Calleros G and Torres-Moran JP. 2013. Insecticide resistance in adult Diaphorina citri Kuwayama from lime orchards in Central West Mexico. Southwestern Entomologist. 38: 579-596. http://doi.org/10.3958/059.038.0404 [ Links ]
Yu K, Moreira-Soares J, Kumar-Mandal M, Wang C, Chanda B, Gifford AN, Fowler JS, Navarre D, Kachroo A and Kachroo P. 2013. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI-1 mediates Azalaic-Acid-induced system immunity. Cell Reports. 3(4):1266-1278. http://doi.org/10.1016/j.celrep.2013.03.030 [ Links ]
Zhang M, Guo Y, Powell C, Doud M, Yang C and Duan Y. 2014. Effective antibiotics against ‘Candidatus Liberibacter asiaticus’ in HLB-affected Citrus plants identified via the graft-based evaluation. Public Library of Science One. 9:11: e111032. https://doi.org/10.1371/journal.pone.0111032 [ Links ]
Zhang M, Powell CA, Guo Y, Doud MS and Duan Y. 2012 A graft-based chemotherapy method for screening effective molecules and rescuing huanglongbing-affected citrus plants. Phytopathology. 102(6): 567-574. https://doi.org/10.1094/PHYTO-09-11-0265 [ Links ]
Received: June 30, 2019; Accepted: August 16, 2019











 text in
text in 



