Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.36 n.1 Texcoco Jan./Apr. 2018
https://doi.org/10.18781/r.mex.fit.1706-2
Scientific articles
Presence of Papaya ringspot virus (PRSV) in weed associated with Carica papaya in Colima, Mexico
1 INIFAP Campo experimental Tecomán. Km 35 Autopista Colima-Manzanillo. C.P. 28100, Tecomán, Colima.
2 Universidad de Colima, Km 40 Autopista Colima-Manzanillo, C.P. 28100, Tecomán, Colima.
3 Colegio de Postgraduados, Campus Campeche. Km. Carretera Haltunchen-Edzna Km. 17.5, C.P. 24450, Sihochac, Champotón, Campeche.
4 CUCBA. Universidad de Guadalajara. Camino Ramón Padilla Sánchez No. 2100 Nextipac, C.P. 45200, Zapopan, Jalisco.
5 TecNM-Instituto Tecnológico de Tlajomulco. Km 10 Carretera a San Miguel de Cuyutlán, C.P. 45640, Tlajomulco de Zúñiga, Jalisco.
Papaya ringspot virus (PRSV) is a Potyvirus of economic importance for papaya crops (Carica papaya L.) whose main symptoms are mosaic, chlorosis and deformation in leaves, as well as ring-like characteristic spots on the fruit. In Mexico, there is no report on weed species associated with this crop that act as reservoir of Potyvirus. In the present study, the presence of PRSV in weeds of commercial papaya plantations was identified in the state of Colima, Mexico. In a sample of 139 papaya plants and 70 weed samples with symptoms of viral infections, PRSV was detected in 40 and 50% for Carica papaya and Cucurbita pepo, respectively, using DAS-ELISA (Double antibody sandwich enzyme-linked immunosorbent assays). Nucleotide sequence analyses of the 350 base pairs fragments of the PRbV NIb region generated by RT-PCR (Reverse Transcription-Polymerase Chain Reaction) allowed the identification of Potyvirus in 40-90% of the total samples of Abutilon abutilastrum and Anoda cristata (Malvaceae), as well as Solanum rostratum (Solanaceaea), which represents a new report on PRSV reservoir weeds associated with papaya crops in Mexico.
Key words: weed; alternate hosts; DAS-ELISA; RT-PCR; Potyvirus
Papaya ringspot virus (PRSV) es un Potyvirus de importancia económica para el cultivo de papayo (Carica papaya L.) produciendo síntomas de mosaico, clorosis y deformación en las hojas, así como manchas características en forma de anillo en el fruto. En México no existen reportes de especies de arvenses asociadas a este cultivo que actúen como reservorio del Potyvirus. En el presente trabajo, se identificó la presencia de PRSV en arvenses de plantaciones comerciales de papayo del estado de Colima, México. En un muestreo de 139 plantas de papayo y 70 arvenses con sintomatología de virosis, se detectó a PRSV en un 40 y 50% para Carica papaya y Cucurbita pepo respectivamente mediante DAS-ELISA (Ensayo por inmunoabsorción ligado a enzimas con doble anticuerpo en sándwich), mientras que el análisis de la secuencia de 350 pares de bases de la región NIb de PRSV generados por RT-PCR (reacción en cadena de la polimerasa con transcriptasa reversa) permitió identificar al Potyvirus en un 40 a 90% del total de muestras de Abutilon abutilastrum y Anoda cristata (Malvaceae), así como Solanum rostratum (Solanaceaea), lo que representa un nuevo reporte de arvenses reservorio de PRSV asociadas a plantaciones de papayo en México.
Palabras clave: maleza; hospederos alternos; DAS-ELISA; RT-PCR; Potyvirus
Papaya (Carica papaya L.) is a tropical herbaceous plant native to Mexico and Central America whose fruit is commercially important. Ranked fifth among exporters worldwide, Mexico produces 951,921 tons of papaya. Colima holds the second place in papaya domestic production with 146,244 ton on an area of nearly 1,800 ha, and the first place with 60% of papaya exports. Papaya cultivation creates around 1,500 direct employments and economic benefits ranging from 300 to 400 million pesos a year (SIAP, 2016).
Plague and diseases are important factors that reduce the quality of papaya fruit for human consumption. Viruses affecting papaya crops cause diseases of economic importance worldwide that pose a serious threaten because they reduce fruit production or even cause the total loss of infested plantations (Navanath et al., 2017). Over 10 viruses affecting papaya have been reported worldwide from which the most important are Papaya leaf distortion mosaic virus (PLDMV), Papaya lethal yellowing virus (PLYV), Papaya mosaic virus (PapMV), Papaya melaira virus (PMeV) and Papaya ringspot virus (PRSV) (Abreu et al., 2015). The latter belongs to the family Potyviridae, genus Potyvirus (Fauquet et al., 2005), it is transmitted by vector aphids in a non-persistent manner, develops symptoms in the form of mosaics, chlorosis, leaf distortion and ring-like spots, and is responsible for crop losses ranging from 50 to 90%, and from 30 to 40% at postharvest (Hernández-Castro et al., 2004; Hernández-Castro et al., 2015).
It has been observed that PRSV’s survival greatly depends on its capacity to spread from host to host and that weeds in papaya crops can serve as PRSV reservoirs. Isolates of this virus belong mainly to two different strains known as PRSV-P (papaya) and PRSV-W (watermelon), where the hosts range of the P strain is limited to plants of the Caricaceae, Cucurbitaceae and Chenopodiaceae families, whereas the isolates of the W strain mainly infect plants of the Cucurbitaceae and Chenopodiaceae families (Chin et al., 2007; Tripathi et al., 2008).
Research has been conducted at international level to identify papaya weeds carrying PRSV. In Brazil, a study was conducted to evaluate natural infection of cucurbitaceous species inside and outside of papaya crops infected with PRSV-P using ELISA and RT-PCR tests (Mansilla et al., 2013). In Florida, the presence of PRSV-W was reported in plant species of the family Cucurbitaceae that damage cucumber, squash, melon and watermelon crops; PRSV-W was also reported as a host of Momordica balsamina, Melothria pendula and Coccinia grandis (Goyal et al., 2012; Tantiwanich et al., 2014). In Jamaica, using DAS-ELISA, it was found that M. charantia is a host for PRSV-P (Chin et al., 2007). In addition to reports from other countries, in Mexico there is not a description of PRSV host weeds associated with papaya crops considering that there is about 30% to 100% of incidence of such virus (Hernández-Castro et al., 2015). This research was aimed at determining the presence of PRSV in some virus-host plant weed species associated with papaya commercial crops in Colima, Mexico, using serological and molecular techniques.
Materials and methods
Plant material
A sampling of papaya plants and weeds was conducted in the state of Colima, Mexico, from March to September 2014, in three plantations at La Cumbre and Los Asmoles, localities from the municipality of Colima, and Camino to Chanchopa from the municipality of Tecomán. Leaves of various weed species showing overall viral symptoms (leaf malformation, chlorosis, mosaic and corrugated leaves) were sampled. Leaves of different papaya plants with PRSV characteristic symptoms were also collected at the same plantations. To identify the gender and species of the collected weeds, dichotomous keys were used according to Fryxell, (1988).
Serological detection of PRSV
The serological detection of PRSV in all leaf samples was performed using DAS-ELISA and the PhatoScreen® (Agdia) kit. The samples were macerated in an extraction shock-absorber and then 100 µL of raw extract were taken and placed in ELISA plates along with the positive and negative controls provided in the kit. The incubations, the enzyme-substrate conjugate, as well as the corresponding washings were conducted following the manufacturer’s instructions, this is, measuring wells at 405 nm. The absorbance value three times higher than that of the absorbance of the negative control was taken as a positive value. Each analysis was carried out in triplicate. Based on those results, a descriptive analysis was performed to determine the percentage of the samples positive to PRSV.
Extraction and purification of RNA
To extract PRSV viral RNA, 0.5 g of leaves were weighed and powdered with liquid nitrogen. Fifty mg of powder were immediately dissolved in 500 μL de TRIZOL® (LifeTechnologies™). The mixture was homogenized and incubated for 5 min at room temperature. It was centrifuged at 12,000 rpm for 10 min at 4 °C, and the aqueous phase was transferred to a new tube adding 100 µL of chloroform; the mixture was vigorously agitated and incubated at room temperature for 3 min. The mixture was centrifuged at 12,000 rpm for 15 min at 4 °C. To the aqueous phase recovered, 250 µL of cold isopropyl alcohol were added, and then centrifuged at 12,000 rpm for 10 min at 4 °C. The tablet obtained in the previous process was washed in 500 µL of 75% ethanol, dried at room temperature and re-suspended in RNA-free water treated with diethyl dicarbonate. The RNA concentration and purity were verified in a NanoDrop spectrophotometer with an absorbance ratio of 260 and 280 nm, while the RNA integrity was verified in 1% agarose gel electrophoresis with 10 mM sodium borate buffer stained with ethidium bromide.
Detection of Potyvirus through reverse transcription polymerase chain reaction (RT-PCR)
The RNA reverse transcription (RT) was performed with the kit “GoScript™ Reverse Transcription System” (Promega A5001. Madison, WI. USA). The RNA was denatured at 70 °C for 10 min while the RT reaction mixture was prepared; the mixture contained 2 µL of ribonuclease inhibitor (Recombinant RNasin), 8 U of AMV reverse transcriptase (0.32 µL), 0.25 µg of random oligonucleotides (0.5 µL) and 5 µL of total RNA (5 ng). The mixture was incubated at room temperature for 10 min and then at 48 °C for 45 min. Immediately afterward it was incubated at 94 °C for 2 min, and finally at 5 °C. The DNAc was amplified using PCR and universal oligonucleotides for Potyvirus as described by Zheng et al. (2010). The last reaction mixture for PCR was prepared in a 25 µL volume containing 17.5 µL of MQ water, 2.5 µL of 10X polymerase buffer, 1 µL of MgCl2 50 mM, 0.5 µL of dNTPs 10 mM, 1 µL of each of the oligonucleotides NIb2F (GTI TGY GTI GAY GAY TTY AAY AA) and NIb3R (TCI ACI ACI GTI GAI GGY TGN CC), 0.5 µL of Taq DNA polymerase (5U/µL) (Invitrogen) and 1 µL of DNAc. The reaction mixture was incubated in a thermocycler under the following conditions: denaturation at 95 °C for 3 min, followed by 35 denaturation cycles at 95 °C for 45 sec, alignment at 45 °C for 45 sec, extension at 72 °C for 45 sec, and a final extension at 72 °C for 5 min. When the experiment ended, the mixture was kept at 4 °C. 12.5 µL of the PCR product was tested by 1% agarose gel electrophoresis using SB (sodium borate, 10 mM) at 100 V. Finally, the amplification products were visualized using a transilluminator with UV light.
Cloning of PCR products
The PCR products were purified using the “PCR Clean-up” (Lamda Biotech) kit, but those showing more than one amplified band were purified using agarose gel and the “Rapid Gel Extraction System” kit (Marligen). Later on, the purified fragments were cloned in the commercial vector “pGEM-T Easy” (Promega) and inserted into competent E. coli cells JM109 (Promega), according to the methodology of Riley et al. (2008).
Secuencing and secuencing analysis
The extraction of plasmidic DNA was performed using the “Pure Link® Quick Plasmid Miniprep” (Invitrogen) kit; the plasmid was then sequenced in a ABI PRISM 310 Genetic Analyzer by the termination method using Big Dye from Applied Biosystems. The bioinformatics analysis was performed using CLC Genomics Workbench version 4. The similarity of the obtained sequences was compared with the sequences reported for PRSV in the NCBI database using the “Basic Local Alignment Search Tool” (BLAST) to determine the origin of the isolates.
Results and discussion
Identifying weeds and alternative hosts for PRSV
The genus of the grass-type weed was identified as Eragrostis spp. Another group of weeds were identified within the Herissantia genus. In this research, all the weed species that gave positive for PRSV were identified according to their morphology and the use of descriptors, according to Fryxell (1988). Papaya plantations showed different phenology and cultural labor to control weeds (Figure 1). The papaya crops at La Cumbre and Chanchopa were being harvested at 13 and 12 months of age, respectively. At La Cumbre there were several papaya plantations that were different ages; the weed samples were collected from a one-month old crop that was at the vegetative development stage. Weeds were managed adequately at the plantations at La Cumbre and Chanchopa, while at the Los Asmoles site there was no weed management; as a result, a great diversity and quantity of weed species sprang up there. This condition represents a source of infection for phytopathogens, because when crops are abandoned, or there are no weed management practices, mixed viral infections become increasingly frequent (Bermúdez-Guzmán et al., 2016). In some cases, weeds are interspersed with plants in the form of tramp crops. However, it is very important to ensure that such weeds are not virus hosts that attract possible vectors like aphids that contribute to the rapid spread of the virus (Kalleshwaraswamy and Kumar, 2008). Overall, the sampled papaya plants showed mosaic, leaf deformation, oily stains in the stem and ring-like spots in fruit, all of them characteristic symptoms of PRSV (Figure 1). The symptoms observed in the field were similar to those reported in papaya plants in several states of Mexico, as well as in Hawaii, Florida, Taiwan and Thailand, to name a few (Noa-Carranza et al., 2006; Tripathi et al., 2008).
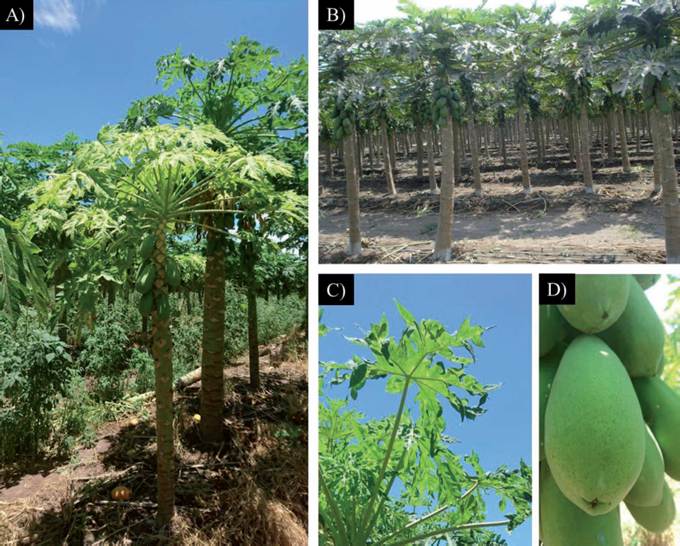
Figure 1 A) Symptoms of PRSV in a papaya plantation where there are abundant interspersed weeds; B) Plantation with full control of weeds; C) Distorted leaves of papaya with chlorosis and mosaic symptoms; D) Fruit with charac-teristic rings caused by PRSV.
Martins et al. (2016) recently collected in Brazil aphid vectors present in 22 weed species in papaya plantations, most of them belonging to the Euphorbiaceae, Commelinaceae, Brassicaceae and Malvaceae families. They also identified one Solanaceae species: Solanum americanum. Chin et al. (2007) reported that Momordica charantia is a PRSV-P carrier; however, Spadotti et al. (2013), after evaluating it through mechanical inoculation and aphid vectors, stated that Momordica charantia is not susceptible to any of the PRSV strains (P and W). In this research, we found a M. charantia specimen from Los Asmoles that appeared healthy and gave negative DAS-ELISA and RT-PCR test results. However, in the future it would be useful to analyze a greater number of samples of this species to determine if M. charantia can act as a reservoir of the virus.
The expression of the multiple viral symptoms in weeds associated with papaya crop in three localities in Colima is shown in Figure 2. The weedsamples that developed symptoms account for 72% of the total sampling, except for Eragrosis spp. and Herissantia spp. Mosaics were observed in plants identified as abutilon (Abutilon abutilastrum), spurred anoda (Anoda cristata) and buffalobur nightshade (Solanum rostratum). The latter species also developed leaf distortion and chlorosis, the same as summer squash (Cucurbita pepo), whose leaf deformation was quite severe. On the other hand, an asymptomatic specimen of Momordica charantia was found in Los Asmoles, which has been reported as host for several viruses; C. pepo, S. rostratum and A. cristata are susceptible to 62, 8 and 1 viruses, respectively; for the Abutilon genus, five viral phytopathogens have been reported (Brunt et al., 1996. When weeds serve as a reservoir of a large number of viruses, this often means that the symptoms observed are the result of the synergic expression of a mixture of viral infections (Zhang et al., 2001; Hii et al., 2002; Gastélum et al., 2007; Bermúdez-Guzmán et al., 2016).
Detection of PRSV
Data on the detection of PRSV through DAS-ELISA and RT-PCR in plants of C. papaya and weeds associated with the crop from three Colima sites are shown in Table 1. From the 130 collected samples, 57 collected at La Cumbre were analyzed using DAS-ELISA and gave positive to PRSV in C. papaya and C. pepo. This result may be explained by the fact that the analyzed samples did not reach the minimum viral concentration since, according to reports, the DAS-ELISA technique does not detect virus in plants if its concentration level is less than 0.1 ng of virus per mL (Salazar, 1996). In this study, up to 50% of positive samples was reported by DAS-ELISA, a percentage highly significant and different from the percentage obtained by Hernández de la Cruz et al. (2007), who detected 2.43% of PVYNTN in potato tubers using the DAS-ELISA technique.
Table 1 Detection of PRSV in weeds associated with papaya crop in three sites in Colima Mexico, using serological and molecular methods.
| Positivas a PRSV (%) | |||||
| Localidad | Muestras colectadas |
Especie | Familia | DAS-ELISA | RT-PCR |
| La Cumbre | 57 | C. papaya | Caricaceae | 40 | 40 |
| A. cristata | Malvaceae | 0 | 21 | ||
| C. pepo | Cucurbitaceae | 50 | 54 | ||
| Eragrosis spp. | Poaceae | 0 | 0 | ||
| S. rostratum | Solanaceae | 0 | 18 | ||
| Los Asmoles | 35 | C. papaya | Caricaceae | 40 | 90 |
| A. abutilastrum | Malvaceae | 0 | 28 | ||
| C. pepo | Cucurbitaceae | 50 | 80 | ||
| M. charantia | Cucurbitaceae | 0 | 0 | ||
| S. rostratum | Solanaceae | 20 | 33 | ||
| Chanchopa | 47 | C. papaya | Caricaceae | 40 | 75 |
| C. pepo | Cucurbitaceae | 0 | 0 | ||
| Herissantia spp. | Malvaceae | 0 | 0 | ||
The BLASTn analysis of the 13 sequences obtained in the present research that correspond to the NIb region of PRSV allowed for the first time the identification of A. abutilastrum, A. cristata and S. rostratum as new natural host species for PRSV. Isolated sequences of C. papaya and C. pepo were also included, but they have been extensively reported as hosts for PRSV (Noa-Carranza et al., 2006; Sreenivasulu and Sai-Gopal, 2010; Goyal et al., 2012; Mansilla et al., 2013). Phylogenetics (Figure 3) was useful to identify three similar sequence groups. A clear grouping is seen according to the plantation zone; for example, it was observed that sample 11 and 12 corresponding to C. pepo and A. abutilastrum, respectively, was the same as the one found in C. papaya corresponding to sequences 9 and 8 from Los Asmoles. This finding clearly confirms that the weed transmits the same virus to papaya plants. Another major finding was the diverse sequences identified for the Colima zone which are markedly different from other isolates, even from the one reported for Veracruz (AY231130) with 92% homology. Such diversity of PRSV isolates has been observed in regions of China due to the high mutation rate of the virus (Zhao et al., 2016), which makes difficult to implement control strategies. As expected, other Potyvirus such as: Japanese yam mosaic potyvirus (JYMV), Potato virus Y (PVY), Bean yellow mosaic virus (BYMV) and Tobacco vein banding mosaic virus (TVBMV) were located farther away in the dendrogram with 70 to 73% identity in the sequence.
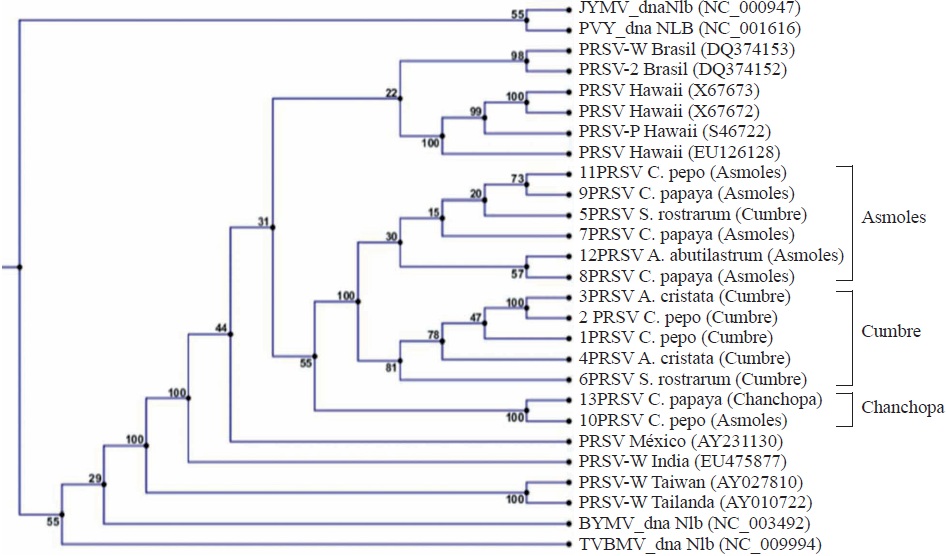
Figure 3 Phylogeny based on the similarity of the sequences of the NIb region of the Papaya ringspot virus (PRSV) genome from isolates collected in Colima, Mexico (1-13) and compared with isolates from Brazil, Hawaii, India, Taiwan, Thailand, and other Potivirus: Japanese yam mosaic potyvirus (JYMV), Potato virus Y (PVY), Bean yellow mosaic virus (BYMV), Tobacco vein banding mosaic virus (TVBMV).
28.15% of papaya plants analyzed in this research developed viral symptoms but they gave negative to PRSV when tested using RT-PCR. This may be due to the fact that C. papaya is susceptible to more than eight phytopathogen viruses such as Papaya apical necrosis virus (PANV), Papaya lethal yellowing virus (PLYV), Papaya Mosaic Virus (PMV), Papaya leaf distortion mosaic virus (PLDMV), among others (Lastra and Quintero, 1981; Brunt et al., 1996), so it may be that the viral symptom may be caused by some of those viruses.
Conclusions
The DAS-ELISA technique allowed the detection of Papaya ringspot virus (PRSV) in C. papaya and any other weed present in papaya crops. However, the number of positive samples was underestimated when they were compared using RT-PCR. This technique was useful to identify the presence of PRSV in weed species associated with papaya crop, such as A. abutilastrum, A. cristata, C. pepo and S. rostratum. PRSV was not detected in M. charantia. This research reports for the first time that A. abutilastrum and A. cristata, belonging to the Malvaceae family, and S. rostratum belonging to the Solanaceae family, are host weed species for PRSV.
Literatura citada
Abreu PMV, Antunes TFS, Magaña-Álvarez A, Pérez-Brito D, Tapia-Tussell R, Ventura JA, Fernandes AAR and Fernandes PMB. 2015. A current overview of the Papaya meleira virus, an unusual plant virus. Viruses 7: 1853-1870. http://dx.doi.org/10.3390/v7041853 [ Links ]
Bermúdez-Guzmán MJ, García-Mariscal KP, Orozco-Santos M, Guzmán-González S, Velázquez-Monreal JJ, García-Preciado JC, Cervantes-Preciado JF y Álvarez-Cilva M. 2016. Enfermedades ocasionadas por virus en caña de azúcar en el Occidente de México. Campo Experimental Tecomán, INIFAP. Folleto técnico No. 13. Tecomán, Col., México. 27p. http://dx.doi.org/10.13140/RG.2.2.15748.73605 [ Links ]
Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L and Zurcher EJ. 1996. Plant Viruses Online: Descriptions and Lists from the VIDE Database. Version: 16th January 1997. Disponible en línea: http://biology.anu.edu.au/Groups/MES/vide/ [ Links ]
Chin M, Ahmad MH and Tennant P. 2007. Momordica charantia is a weed host reservoir for Papaya ringspot virus Type P in Jamaica. Plant Disease. 91: 1518. http://dx.doi.org/10.1094/PDIS-91-11-1518A [ Links ]
Fauquet CM, Mayo MA, Maniloff J, Desselbelguer U and Ball LA. 2005. Virus taxonomy: the eighth report of the International Committee on Taxonomy of Viruses. 1st Edition. Academic Press Elsevier. Amsterdam, Holanda. 1162p. http://dx.doi.org/10.1007/978-3-7091-6607-9 [ Links ]
Fryxell, P. A. Malvaceae of Mexico. Syst. Bot. Monogr. 25: 1-522. 1988. Disponible en línea: http://www1.inecol.edu.mx/publicaciones/resumeness/FLOBA/Flora%2016.pdf [ Links ]
Gastélum RB, Magallanes-Tapia MA, Méndez-Lozano J, Huet H, Trigueros-Salmerón JA y Longoria-Espinosa RM. 2007. Detección del virus mosaico amarillo de la calabaza zucchini (ZYMV) y su coinfección con otros virus en cucurbitáceas cultivadas y plantas silvestres en el valle del fuerte, Sinaloa, México. Revista Mexicana de Fitopatología. 25:95-101. Disponible en línea: http://www.redalyc.org/pdf/612/61225201.pdf [ Links ]
Goyal G, Gill HK and McSorley R. 2012. Common weed hosts of insect-transmitted viruses of Florida vegetable crops. Disponible en línea: http://edis.ifas.ufl.edu/in931 [ Links ]
Hernández-Castro E, Villanueva-Jiménez JA, Mosqueda- Vázquez R y Mora-Aguilera JA. 2004. Efecto de la erradicación de plantas enfermas por el PRSV-P en un sistema de manejo integrado del papayo (Carica papaya L.) en Veracruz, México. Revista Mexicana de Fitopatología 22: 382-388. Disponible en línea: http://www.redalyc.org/articulo.oa?id=61222311 [ Links ]
Hernández-Castro E, Damian-Nava A, Mora-Aguilera A, Villanueva-Jimenez JA, Vargas-Álvarez D and Palemón-Alberto F. 2015. Incidence of the Papaya Ringspot Virus (PRSV-p) and Management in the State of Guerrero, Mexico. Pp:119-127 In: S. Dimitrov Todorov and I. Vitanova Ivanova (eds.) Tropical Fruits. Chapter 7. Nova Science Publishers, Inc. http://dx.doi.org/10.13140/RG.2.1.3437.5203 [ Links ]
Hernández de la Cruz M, Gómez-Leyva JF, López-Muraira IG, Dimas-Estrada MS, Andrade-González I y Ireta-Moreno J. 2007. Detección serológica y molecular del virus PVYN y su variante PVYNTN en papa (Solanum tuberosum L.) y hospedantes alternos en Tapalpa, México. Revista Mexicana de Fitopatología. 25: 167-172. Disponible en línea: http://www.redalyc.org/pdf/612/61225211.pdf [ Links ]
Hii G, Pennington R, Hartson S, Taylor CD, Lartey R, Williams A, Lewis D and Melcher U. 2002. Isolate-specific synergy in disease symptoms between cauliflower mosaic and turnip vein-clearing viruses. Archives Virology 147: 1371-1384. http://dx.doi.org/10.1007/s00705-002-0812-8 [ Links ]
Kalleshwaraswamy CM and Kumar NK. 2008. Transmission efficiency of Papaya ringspot virus by three aphid species. Phytopathology. 98: 541-546. Disponible en línea: http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO-98-5-0541 [ Links ]
Lastra R and Quintero E. 1981. Papaya apical necrosis, new diseases associated with a Rhabdovirus. Plant Disease 65: 439-440. http://dx.doi.org/10.1094/PD-65-439 [ Links ]
Mansilla PJ, Moreira AG, Mello APOA, Rezende JAM, Ventura JA, Yuki VA and Levatti FJ. 2013. Importance of cucurbits in the epidemiology of Papaya ringspot virus type P. Plant Pathology 62: 571-577. http://dx.doi.org/10.1111/j.1365-3059.2012.02677.x [ Links ]
Martins DS, Ventura JA, Paula RCAL, Fornazier, MJ, Rezende, JAM, Culik MP, Ferreira PSF, Peronti ALBG, Carvalho RCZ and Sousa-Silva CR. 2016. Aphid vectors of Papaya ringspot virus and their weed host in orchards in the major papaya producing and exporting region of Brazil. Crop Protection 90: 191-196. https://doi.org/10.1016/j.cropro.2016.08.030 [ Links ]
Navanath MD, Anitha P, Gahukar SJ, Akhare AA. 2017. Biological indexing of Papaya ring spot virus (PRSV) in Carica papaya. International Journal of Agriculture 9: 3728-3730. Disponible en línea: https://bioinfopublication.org/files/articles/9_4_7_IJAS.pdf [ Links ]
Noa-Carranza JC, González de León D, Ruiz-Castro BS, Piñero D and Silva-Rosales L. 2006. Distribution of Papaya ringspot virus and Papaya mosaic virus in papaya plants (Carica papaya) in Mexico. Plant Disease. 90: 1004-1011. https://doi.org/10.1094/PD-90-1004 [ Links ]
Riley SP, Woodman ME and Stevenson B. 2008. Culture of Escherichia coli and related bacteria. Current Protocols Essential Laboratory Techniques. Wiley. U 4.2.168p. https://doi.org/10.1002/9780470089941.et0402s00 [ Links ]
Salazar LF. 1996. Potato viruses and their control. 1st Edition. International Potato Center (CIP). Lima, Perú. 214p. Disponible en línea: https://cipotato.org/publications/potato-viruses-and-their-control/ [ Links ]
SIAP 2016. Secretaría de Agricultura, Ganadería, Rural, Pesca y Alimentación. Servicio de Información Agroalimentaria y Pesquera. Disponible en línea: www.siap.gob.mx/index [ Links ]
Spadotti DMA, Buriolla JE, Rezende JAM and Souza VC. 2013. The wild type of Momordica charantia is not infected by potyviruses that cause disease in papaya and cucurbit crops. Tropical Plant Pathology 38: 447-451. http://dx.doi.org/10.1590/S1982-56762013005000029 [ Links ]
Sreenivasulu M and Sai-Gopal DVR. 2010. Development of recombinant coat protein antibody based IC-RT-PCR and comparison of its sensitivity with other immunoassays for the detection of Papaya ringspot virus isolates from India. The Plant Pathology Journal. 26: 25-31. http://dx.doi.org/10.5423/PPJ.2010.26.1.025 [ Links ]
Tantiwanich Y, Baker CA, Turechek WW and Adkins S. 2014. Detection of Papaya ringspot virus type W infecting the cucurbit weed Cucumis melo var. dudaim in Florida. Plant Health Progress 15: 29-30. http://dx.doi.org/10.1094/PHP-BR-13-0126 [ Links ]
Tripathi S, Suzuki JY, Ferreira SA and Gonsalves D. 2008. Papaya ringspot virus-P: characteristics, pathogenicity, sequence variabality and control. Molecular Plant Pathology. 9: 269-280. http://dx.doi.org/10.1111/j.1364-3703.2008.00467.x [ Links ]
Zhao H, Zong RJ, Zhang YL, Zhu YJ, Zeng HC, Kong H, McCafferty H, Guo AP, and Peng M. 2016. Geographical and Genetic Divergence Among Papaya ringspot virus Populations Within Hainan Province, China, Virology 106:937-944. https://doi.org/10.1094/PHYTO-05-15-0111-R [ Links ]
Zhang XS, Holt J and Colvin J. 2001. Synergism between plant viruses: a mathematical analysis of the epidemiological implications. Plant Pathology 50: 732-746. http://dx.doi.org/10.1046/j.1365-3059.2001.00613.x [ Links ]
Zheng L, Rodoni BC, Gibbs MJ and Gibbs AJ. 2010. A novel pair of universal primers for the detection of potyviruses. Plant Pathology. 59: 211-220. http://dx.doi.org/10.1111/j.1365-3059.2009.02201.x [ Links ]
Received: June 21, 2017; Accepted: August 31, 2017











 text in
text in 



