Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.35 n.1 Texcoco Jan. 2017
https://doi.org/10.18781/r.mex.fit.1605-4
Scientific articles
Morphological and molecular characterization of native isolates of Trichoderma and its potential biocontrol against Phytophthora infestans
1Instituto de Ciencias Agropecuarias y Rurales (ICAR), Universidad Autónoma del Estado de México, Campus Cerrillo Piedras Blancas, Carretera a Tlachaloya s/n, Toluca, Estado de México, CP 50090.
2Instituto Hispano-Luso de Investigaciones Agrarias (CIALE), Universidad de Salamanca, Campus de Villamayor C/ Del Duero, 12; 37185 Villamayor (Salamanca), España.
3Facultad de Ciencias, Universidad Autónoma del Estado de México, Campus Cerrillo Piedras Blancas, Carretera Toluca-Atlacomulco, Estado de México, CP 50090, México.
4Laboratorio de Agroecología, Instituto de Investigaciones en Ecosistemas y Sustentabilidad (IIES), Universidad Nacional Autónoma de México. Antigua Carretera a Pátzcuaro 8701 Col. Ex-Hacienda de San José de la Huerta C.P. 58190, Morelia, Michoacán, México.
This paper presents the morphological and molecular characterization of 10 native isolates of Trichoderma (TL2, TL4, TL5, TL6, TX7, TX8, TT6, TF8, TF10 and TJ6) and their phylogenetic relation, as well as its biocontrol ability against Phytophthora infestans. The results of the molecular characterization by amplifying the ITS1ITS4 region of nucle1 and TJ6). The phylogenetic analysis by maximum parsimony showed a close relation among the isolates of these two groups. Biocontrol tests between the antagonist and the pathogen showed significant differences (p<0.05). Strain TX8 had the highest inhibition percentage (98 %), while TL4 recorded the lowest percentage (49 %). The isolates of T. harzianum (TL5, TL6, TF10 y TJ6) showed percentages between 5565 %.
Key words: T. asperellum; T. harzianum; antagonism; potato; Solanum tuberosum
El presente trabajo muestra la caracterización morfológica y molecular de 10 cepas nativas de Trichoderma (TL2, TL4, TL5, TL6, TX7, TX8, TT6, TF8, TF10 y TJ6) y su relación filogenética, así como su capacidad de biocontrol contra Phytophthora infestans. Los resultados de la caracterización molecular al amplificar la región ITS1-ITS4 del ADN ribosómico nuclear y un fragmento del gen tef1α mostraron que de las 10 cepas, seis corresponden a Trichoderma asperellum (TL2, TL4, TX7, TX8, TT6 y TF8) y cuatro a T. harzianum/Hypocrea lixii (TL5, TL6, TF10 y TJ6). El análisis filogenético por máxima parsimonia demostró que existe una estrecha relación entre las cepas de estos dos grupos. Las pruebas in vitro de biocontrol entre el antagonista y el patógeno indicaron diferencias significativas (p<0.05). La cepa TX8 registró el mayor porcentaje (98 %) de inhibición, mientras que TL4 fue la de menor porcentaje (49 %). Las cuatro cepas de H. lixii (TL5, TL6, TF10 y TJ6) presentaron capacidad de inhibición media (55-66 %).
Palabras clave: T. asperellum; H. lixii; antagonismo; papa; Solanum tuberosum
The potato crop (Solanum tuberosum) is of great economic and agricultural importance, since potato is the third most consumed food in the world (FAO, 2014). Potato production in Mexican fields contributes 0.5 % of the worldwide supply (SIAP, 2014). The municipality of Zinacantepec, State of Mexico, recorded a production of 17,535 t in 2014, equivalent to $115,239.06 (SIAP, 2014). Potato yield is affected by the presence of phytopathogenic agents such as he oomycete Phytophthora infestans (Mont) De Bary, which causes a disease known as “late blight” or potato blight (Kerroum et al., 2015). This oomycete causes leaf lesions which take from three to seven days to develop and become epidemics (Rekanović et al., 2011). In recent years, this pathogen has caused significant economic losses in Zinacantepec (López et al., 2013). Chemical control is the most used method; it is successfully applied to seed, leaves and soil, but has residual effects, which causes chemical products to accumulate on water bodies, soil, plants and animals (Miller et al., 2000; Aceves et al., 2008).
Hence, it is important to search for ecological strategies for biological control, including the use of microorganisms that help reduce the presence of pathogens (Aceves et al., 2008). Some species of the Trichoderma genus are known to act as biocontrol agents with broad spectrum of action against pathogens of agricultural crops (Schuster y Schmoll, 2010). This genus uses mechanisms such as: competition, mycoparasitism and antibiosis (Monte, 2001; Harman et al., 2004; Bailey et al., 2008). Commercial formulations containing various species of Trichoderma (T. harzianum, T. viride and T. atroviride) used in the field (Lorito et al., 2010; Mazyar et al., 2010) usually belong to a different region than that where they are applied, which results in ineffective control (Harman et al., 2010). To this regard, Altieri (1999) recommends the use of native species adapted to the ecological conditions of the region where the control is carried out to ensure a successful biocontrol. However, some companies require isolates with biocontrol ability and adaptation to different regions and against diverse pathogenic agents (PHC, 2016).
In the biological control, it is important to assess the antagonistic potential of the isolates in vitro and in plants and carry out a taxonomic characterization, in order to find out what the requirements are for the species to grow and, based on this information, establish a selection criteria to use them as a biocontrol agents, since not all the species of Trichoderma have such ability (Munir et al., 2013). Conventional characterization of Trichoderma species based on morphological characteristics is not enough due to the plasticity of their traits, so it is important to conduct molecular studies to make a more accurate taxonomical diagnosis (Hermosa et al., 2000). Phylogenetic analyses are a tool that supports and validates molecular studies through evolutionary evidence that can reveal convergent or divergent relations among species (Morrone, 2001).
In previous studies (García-Núñez et al., 2012), 10 native isolates of Trichoderma from horticultural soils of the Toluca Valley, Mexico —some of them used in in vitro antagonism tests— showed high potential for the control of Sclerotinia sp., the causing agent of lettuce soft rot. The isolates were not identified at molecular level, so it is important to know to which species they belong to, as well as their potential biocontrol effect on other pathogens. For example, it has been shown that isolates of Trichoderma asperellum are fungi antagonists and phytopathogenic oomycetes (Taghdi et al., 2015). For this reason, the present study was aimed to characterize 10 native isolates of Trichoderma using conventional, molecular and phylogenetic techniques, and assess their biocontrol potential against Phytophthora infestans, the causing agent of late blight in potato crops.
Materials and methods
Study area and origin of the isolates
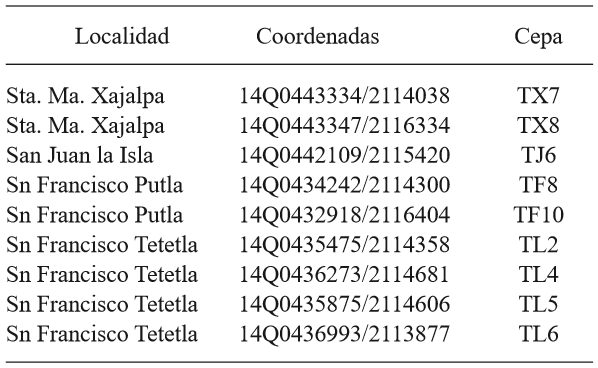
The 10 native isolates of Trichoderma (TL2, TL4, TL5, TL6, TX7, TX8, TT6, TF8, TF10 and TJ6) used in this research were isolated in a previous study (García-Núñez et al., 2012). The isolation points are located in areas of the Toluca Valley horticultural zone (Table 1).
Table 1. Geographic location of the isolates of Trichoderma in the horticultural zone of the Toluca Valley.
One strain of Phytophthora infestans was isolated from a potato crop plot in La Peñuela, a location from the municipality of Zinacantepec, State of Mexico. For this purpose, leaves with late blight signs and symptoms were selected; that is, necrotic stalk and petioles tissue showing dark spots, as well as the presence of white mycelium with gray sporulation on the backside of leaves. The sample was placed on wet paper and taken to the lab, where it was placed on sterilized filter paper and then on Petri dishes with wet cotton to create wet chambers, which were then maintained at room temperature during seven days to foster the presence of mycelium and sporulation. When there was enough mycelium, an evaluation was performed using Sutton’s (1980) technique, described as follows: an active sporulated mycelium was taken with an inoculation loop and put in a Petri dish using a Potato Dextrose Agar (PDA, Bioxon) culture medium. The mycelium was incubated at 22±2°C during 12 days. From this sample, a monosporic isolate was obtained applying the tip-of-hyphae technique. Using the pure strain of P. infestans, tests using Koch’s postulates were conducted to validate its pathogenesis.
The isolates are safeguarded at the Institute of Agricultural and Rural Sciences (ICAR) of the Autonomous University of the State of Mexico.
Macroscopic morphological characterization of isolates
After obtaining pure isolates of Trichoderma and P. infestans, they were morphologically characterized. Isolates of Trichoderma were identified using the guide by Barnett and Hunter (1972), considering the following macroscopic morphological characteristics: mycelium texture, concentric rings development and conidia pigmentation. The strain of P. infestans was characterized using the guide of Erwin and Robeiros (1996) and considering mycelium texture and type, as well as spore pigmentation.
Microscopic morphological characterization
Culture chambers in Petri dishes were built for each strain, according to the modified method by Harris (1986) as follows: a glass triangle containing a slide with a 15 mm in diameter PDA medium disk (Bioxon) was used to inoculate a spore loop of the strain of Trichoderma. The sample was covered with a slide, and the Petri dish was capped and incubated at 25 °C during seven days. Then, the slide containing the active mycelium and isolate spores were removed and placed on a slide; the slide was stained with a drop of cotton blue. Observations were made under an optical microscope (Image Motic Plus 2.0) connected to a digital camera. The images were captured under a target of 100X using immersion oil. For Trichoderma, the description was based on the following characteristics: shape and number of conidia, phialides and conidiophores; this was performed according to the Barnett and Hunter (1972) identification guide. For P. infestans, the morphological characteristics taken into account were: hyphae type; sporangia shape and size; type of papilla or absence of chlamydospores, according to recommendations for oomycetes (Erwin and Robeiro, 1996).
Molecular characterization of Trichoderma
Obtaining mycelium. Petri dishes with a PDA (Bioxon) culture medium and a 76 mm in diameter sterilized cellulose circular membrane (Sigma) were inoculated with 100 μL of a 1X108 spore suspension per mL of each strain. The dishes were incubated at 25 °C during three days. The mycelium recovered from the membrane by scratching with a scratcher under sterility conditions was frozen and lyophilized. The extraction of genomic DNA of the fungi was performed according to the technique described by Hermosa et al. (2000).
Polymerase chain reaction (PCR). The ITS1ITS4 region, nuclear ribosomal DNA zone, was amplified as follows: the commercial primers ITS1f (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4r (5’-TCCTCCGCTTATTGATATGC-3’), with which an amplification of 600 bp was expected. Each reaction was prepared with a final volume of 50 µL containing 5 µL of Buffer + MgCl2 (50 mM), 0.3 µL dNTPs (2 mM/μL), 2 µL of primer (10 µM), 1 µL of Taq polymerase (5U); 1.5 µL of genomic ADN; 38.2 µL of miliQ of water. The reaction was conducted in a thermocycler (MJ Research, Inc.) under the following program: first stage: initial denaturation at 94 °C during 3 min; second stage; 35 cycles each, starting with denaturation at 94 °C for 1 min, followed by alignment at 53 °C for 1 min 30 s, and extension to 72 °C for 2 min; third stage: final extension to 72 °C for 7 min. The product was kept at 4 °C. The DNA fragments obtained from the amplification (3 µL), mixed with 2 μL of charge buffer, were isolated in a mixture of agarose gel (0.8 %) and 3 µL of ethidium bromide. At this stage, five μL of molecular weight marker of 100 bp (Gene ruler, Thermo Scientific) were used. Running conditions for the electrophoresis camera were 81 volts during 30 min. The DNA fragments were observed using a UV light photo documenter.
The PCR fragments obtained were purified according to the gel extraction (NucleoSpin Extract II, Macherey-Nagel) clean-up kit protocol. The DNA quality and concentration was evaluated in a Nanodrop. The DNA bands were observed in 1 % agarose gel. The same procedure was followed to amplify a fragment of the translation elongation factor 1 alpha (tef1α) gen with the primers EF1278f 5’-CATCGAGAAGTTCGAGAAGG-3’ (Chaverri and Samuels, 2003) and TEF1LLEr 5’-AACTTGCAGGCAATGTGG-3’ (Jaklistch et al., 2005). The amplification was performed under the following conditions: initial denaturation at 94 °C for 3 min, 35 denaturation cycles at 94 °C 30 s, alignment at 58°C for 1 min, extension at 72 °C for 1 min, final extension at 72 °C for 7 min.
Sample preparation for sequencing. Eppendorf tubes with 3 µl of purified PCR product (50 ngµL-1), 3 µL of primer (1µM) and 2 µl of miliQ water. The samples were sent to the Sequencing Department of the University of Salamanca, Spain.
Sequences analysis in genomic databases. The sequences obtained were analyzed using FinchTV version 4.1 and DNAstar in order to obtain consensus sequences. Then, a BLAST alignment analysis in the Genbank database was performed. Phylogenetic analysis. To identify the phylogenetic relation of the isolates, external groups to Trichoderma viride and Trichoderma longibrachiatum (Mohammad et al., 2014) were used. Both sequences were aligned using BioEdit Sequence Alignment ver. 7.05 (Hall, 2004); the phylogenetic relations were established using the heuristic method in the parsimony analysis with the program Past version 207 (Harmer et al., 2001). The robustness of the internal branches was assessed with 1000 bootstrap replications.
Antagonism tests
In vitro confrontation tests were performed on 10 native isolates of Trichoderma and Phytophthora infestans, using a modified version of the method of Larralde et al., (2008), as follows: on Petri dishes containing PDA, antagonists and pathogens were confronted by placing a 5 mm diameter circle of active mycelium in face to face on each dish, separated by approximately 5 cm; they were incubated at 25±2 °C for seven days. This test was carried out in batch (in triplicate) using three repetitions of each strain of Trichoderma. The inhibition percentage of their radial growth was monitored every 24 hours using the following formula: PIRG=[(R1-R2)/R1] x100, where R1 is the radial growth of the non-confronted strain of P. infestans, and R2 is the radial growth of Trichoderma confronted against P. infestans. Confrontation test were performed according to a randomized experimental design for each of the 10 native isolates of Trichoderma confronted against P. infestans. The potential of each strain of Trichoderma as a biocontrol agent of P. infestans infection was evaluated by variance analysis and multiple media comparison (Tukey, p<0.05), using the Statistical Analysis System (SAS) software version 9.
Results and Discussion
Macroscopic morphological characterization of Trichoderma and P. infestans
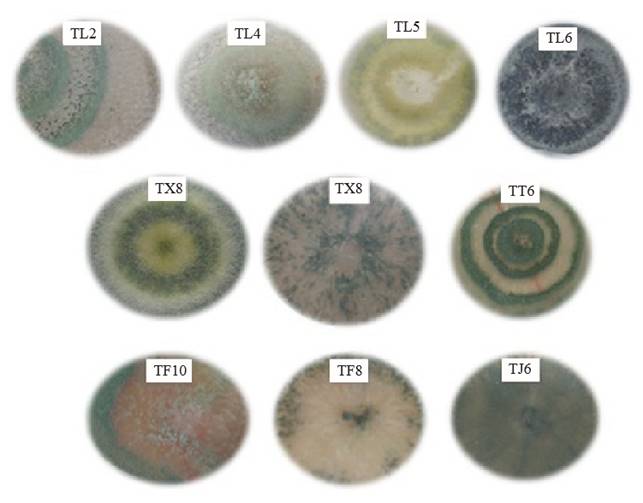
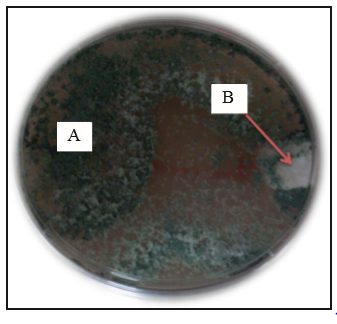
Except for strain TF10, the native isolates of Trichoderma developed abundant fluffy mycelium (Figure 1). Isolates TL2, TL4, TX8 y TT6 developed from two to three well defined concentric white (mycelium) and green (conidia) rings. Strain TL5 developed yellow pigmentation and concentric rings. Meanwhile, no rings were observed in isolates TX7, TF10, TL6 TF8 y TJ6, and the mycelium spread evenly on the dishes. Conidial pigmentation of isolates ranged from light green to dark green. Although the isolates came from the same location, they developed different pigmentation, as in the case of those from San Francisco Tetetla, where TL2, TL4, TL5 and TL6 developed green pigmentation with different tones. The shape and quantity of conidia observed on the dishes may have been caused by factors contributing to a stress environment, such as: light exposure, lack of nutrients or pH changes (Osorio-Concepción et al., 2013). The shape and pigmentation of the mycelium observed in the native isolates are related to the Trichoderma genus, according to Barnett and Hunter (1972) taxonomic identification criteria. These characteristics differ among native isolates and none of them resembles each other; for this reason, it was difficult to group them and identify the species based on these data.
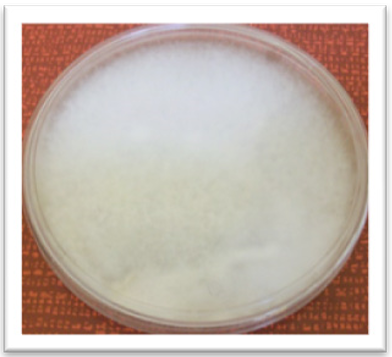
The strain of P. infestans developed abundant white fluffy mycelium with beige tones caused by the presence of sporangia. No concentric ring development was observed (Figure 2).
Microscopic morphological characterization of Trichoderma and P. infestans
P. infestans showed 5-8 µm in diameter hyaline and ramified hyphae, long sporangiosphores with 5 µm pedicel, and ellipsoidal sporangia (28.2 ± 11 X 17.6 ± 1.0 µm) with aged papillae (Figure 3). These results are in agreement with the characteristics reported by Erwin and Robeiro (1996) for P. infestans. They also agree with the description of P. infestans isolates reported by López-Orona et al. (2013).
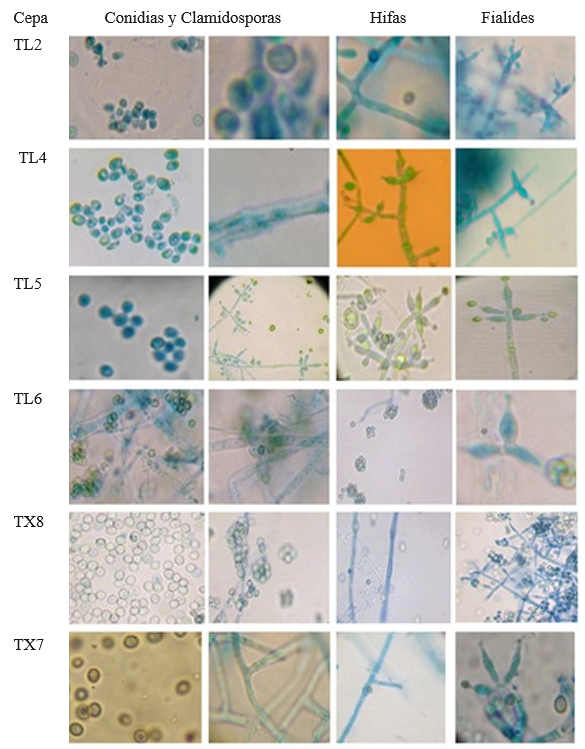
The results from microscopic observations of Trichoderma showed that TF8, TX7, TX8, TT6 and TL2 developed slightly ovoid conidia arranged in rosette-like clusters of 4-6, abundant and spherical chlamydospores, apical phialides arranged in the form of transverse whorls in groups of 3-4, fine thorny ornamentation and ramified conidiophores (Figure 4). These characteristics are in agreement with those reported by Samuels et al., (2010). Strain TL4 developed ovoid conidia and phialides in pairs or groups of three all along the conidiophore that were wider at the center than at the base; based on these characteristics, it can be classified as T. asperelloides. Isolates TL5, TL6, TF10 and TJ6 showed hyaline hyphae, long ramified conidiophores, phialides wide at the center, long at the base and grouped in a pyramid, smooth-walled globose conidia and many terminal chlamydospores; these characteristics show a similarity to Hypocrea (Chaverri and Samuels, 2002; Druzhinina y Kubicek, 2005; Ortiz-García et al., 2015). Since the macroscopic morphological characteristics vary, they do not allow discriminating among species, which highlights the importance of the molecular characterization to taxonomically classify the isolates, given that a diagnosis at the species level based on morphological traits may result in a wrong diagnosis, as in the studies conducted by Hermosa et al., (2000), who performed molecular characterization of isolates from a collection that had been morphologically classified as T. longibranchiatum but that was actually T. asperellum.
Molecular characterization
The genomic DNA amplification of the isolates using primers ITS1 and ITS4 was of 600 bp (Figure 5). The genomic DNA amplification of isolates with the primers pair tef1 278f and tef1ller was 1200 bp (Figure 6). The results from GenBank through a BLAST analysis of the amplified sequences of the two primer pairs showed that from the 10 isolates, six (TL2, TL4, TF8, TT6, TX7 y TX8) were 99 % identical to T. asperellum. The other four isolates (TJ6, TL5, TL6 y TF10) were 99 % identical to T. harzianum a anamorph of Hypocrea lixii (Table 2). Due to the ITS variability, it is sometimes difficult to discriminate among species, so a fragment of the tef1α gen was amplified, as this helped to confirm the taxonomy of the native isolates. This result agree with the findings reported by Druzhinina and Kubiceck (2005), and Kredics et al. (2014), who stated that these markers are essential to characterize Trichoderma/Hypocrea. Also, studies conducted by Ortiz-García et al. (2015) show that ITS sequences are useful to successfully identify Trichoderma/Hypocrea isolates.
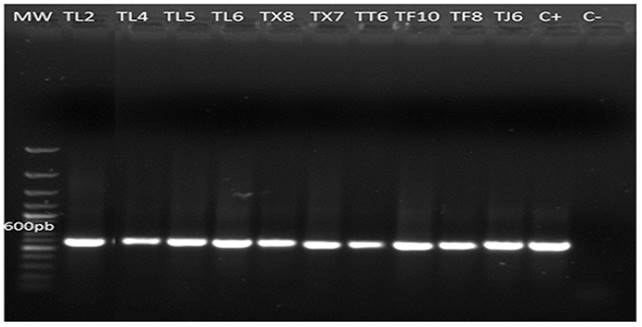
Figure 5. PCR amplification products from genomic DNA of 10 native strains of Trichoderma (TL2, TL4, TL5, TL6, TX8, TX7, TT6, TF10, TF8 and TJ6) at 600bp with primers ITS1, ITS4. MW= molecular weight marker (Gene ruler 100bp plus DNA ladder Thermo Scientific). C+ = Positive control (T. asperellum). C- = Negative control. 0.8 % agarose gel.
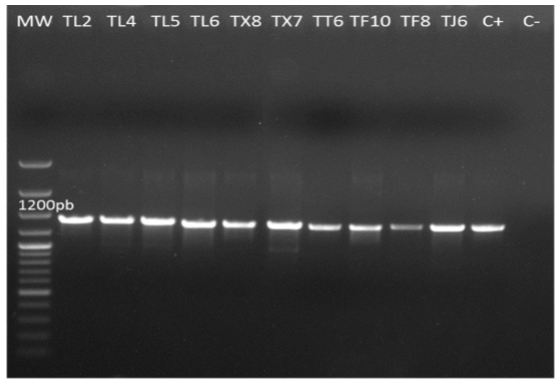
Figure 6. PCR amplification products from genomic DNA of native isolates of Trichoderma (TL2, TL4, TL5, TL6, TX8, TX7, TT6, TF10, TF8 and TJ6) at 1200bp with primers tef1α 278f and tef1ller. MW= molecular weight marker (Gene ruler 100bp plus DNA ladder Thermo Scientific). C+ = Positive control (T. asperellum). C- = Negative control. 0.8 % agarose gel.
Table 2. Trichoderma/Hypocrea species found in the Toluca Valley, Mexico, horticultural zone, identified through amplification of internal transcribed spacers (ITS) and a fragment of the gen translation elongation factor 1 alpha (tef1α).
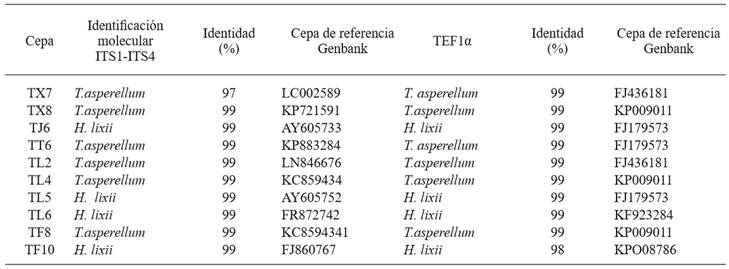
The results obtained show that TL2, TF8, TT6, TX7 and TX8 identified by morphological traits as T. asperellum actually belong to this species. Strain TL4, which was initially classified as T. asperelloides, was reclassified as T. asperellum based on molecular studies. These results show that the molecular identification supports the morphological description and that these elements lead to an accurate and reliable taxonomic diagnosis. On the other hand, these results are similar to those obtained by Guigón et al., (2010), who reported the presence of isolates of T. asperellum in horticultural crops. Isolates TJ6, TL5, TL6 y TF10, which were initially classified as Hypocrea/Trichoderma, were taxonomically identified as H. lixii/T.harzianum through molecular analysis.
The results obtained from characterization of native isolates from the Toluca Valley horticultural zone are significant, since T. harzianum/H. lixii and T. asperellum are species that had not been reported before in this region, and they are important as biological control agents in horticultural crops (Monte, 2001; Harman et al., 2004; Woo et al., 2006.
Phylogenetic analysis
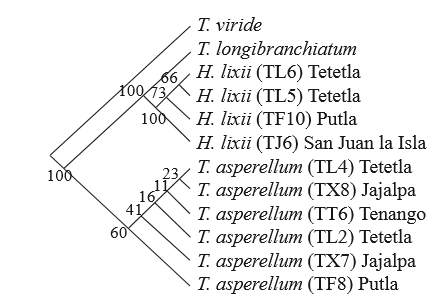
In this case, we performed a heuristic analysis and obtained 82 phylogenetic trees; they were equally parsimonious with a consistency index of 0.97 and 0.99 retention index, which suggests that they were well supported (Figure 7). For statistical bootstraps to obtain branch support values, 1000 replications were used, and values of 60 and 100% were considered as indicators of statistically strong branches. This is important as it confirms that the analyzed groups relate to the expected species, which is similar to the results reported by Sadfi-Zouaoui et al. (2009). The phylogenetic trees show that the most frequently found species was T. asperellum with a wider distribution in the study zone, since the isolates belong to four of the five sampled locations (Figure 7). T. asperellum was found in Tenango del Valle, this finding is being in agreement with studies conducted by Osorio et al., (2005), who reported the presence of T. harzianum and T. lignorum. However, the study was limited only to morphological characterization, and this may have led to a wrong determination. Therefore, it is important to rescue native species with potential as biocontrol agents against plant pathogens such as T. asperellum and T. harzianum/H. lixii. The latter was found in San Francisco Putla (TF10), San Francisco Tetetla (TL5, TL6) and San Juan la Isla (TJ6). It seems that the geographic and environmental conditions established this distribution pattern because of the geographic proximity of the three locations. This was shown by the phylogenetic analysis, replacing taxa by their locations, where a close relation among the species found in the three locations was observed. It is also important to mention that although the 10 isolates belong to the two species, in the tree they are individually grouped, which agrees with the results of the highly diverse macroscopic morphological description. Though their pigmentation and mycelia growth differ from each other, they belong to the same species.
Antagonism tests
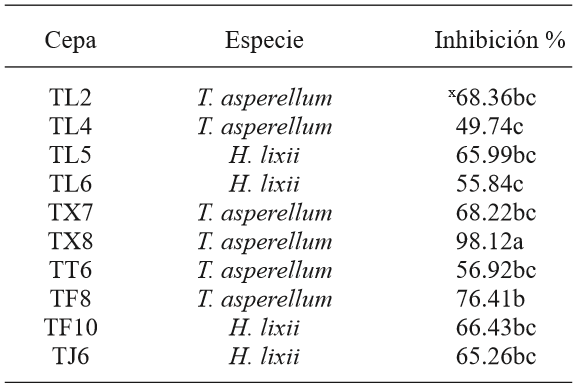
The results from antagonism tests between the 10 native Trichoderma and Phytophthora infestans isolates showed significant differences (p<0.05). In the case of T. asperellum, TX8 and TF8 had the highest percentages of biocontrol of the pathogen (98 and 76 %, respectively) (Figure 8), while TL4 had the lowest value (49 %). Despite of this, we can affirm that T. asperellum showed a higher biocontrol of P. infestans. The variation of these percentages is similar to the in vitro studies conducted by Kerroum et al. (2015), who reported biocontrol values of T. harzianum against P. infestans between 56 to 86 %. The biocontrol percentages obtained in this study are similar to those reported by Al-Mughrabi (2008), where the T. atroviride strain evaluated in vitro showed 92% of control of P. infestans. Furthermore, they exceeded the results obtained by Debebe et al., (2011), where the minimum value of biocontrol of T. viride on the oomycete was 36.7 %. On the other hand, despite T. harzianum/H. lixii has the lowest percentages of biocontrol, these are higher than 50%. These results are in agreement with those obtained by Santos et al. (2013), who reported higher control percentages of T. asperellum compared to T. harzianum/H. lixii in antagonism studies against Colletotrichum gloeosporoides, which causes mango anthracnose.
The results obtained in this study differ from those obtained by García-Núñez et al. (2012) using the same isolates, where the highest inhibition values against Sclerotinia sp belong to isolates TX8 and TL4 of T. asperellum, with 88% and 86%, respectively (Table 3). However, in this study, isolate TL4 (T. asperellum) had the lowest inhibition value against P. infestans (49 %). TF10 (H. lixii) showed similar values with 80 % of control of Sclerotinia sp. and 66 % of P. infestans. Isolates TF8, TX7, TL2, TT6, TJ6, TL5 and TL6 had the lower percentages of biocontrol of Sclerotinia sp., but not for P. infestans. These results agree with those obtained by Matroudi et al, (2013), who reported Trichoderma species with little biocontrol effect of Sclerotinia sclerotiorum, which do not exceed 80%. Also, Aceves et al., (2009) reported Trichoderma species with 5.35 % to 62.9 % of biocontrol. Based on this, the results are interesting, considering that P. infestans is an aggressive pathogen able to mutate easily (López et al., 2013); however, we found T. asperellum isolates with potential to control such oomycete. The isolates of T. harzianum/H. lixii with potential higher than 50% can be strengthened to perform a more effective biocontrol and use them in in vivo tests in further stages, with more virulent isolates.
Conclusions
The morphological and molecular characterization, as well as the phylogenetic analysis performed, provided a reliable diagnosis to identify isolates of Trichoderma isolated in the horticultural zone of the Toluca Valley, Mexico, as Trichoderma asperellum and T. harzianum/Hypocrea lixii. These species are considered to have biological activity of wide action spectrum on crop plant pathogens. The phylogenetic analysis shows the existing relation among the geographic distribution of the two species, where T. asperellum was more frequently found. Strain TX8 identified as T. asperellum, due to its high percentage of biocontrol of P. infestans and Sclerotinia sp., can be a candidate to control these plant pathogens. It can be an agroecological alternative to control the late blight disease in the State of Mexico, given that it is a native strain and may have greater chances of adaptability and success compared to isolates currently available in the market. For this reason, we recommend to conduct studies at greenhouse level and then in the field, including different application doses and methods. The isolates identified can be also be evaluated as biocontrol agents of other pathogens in further studies.
Acknowledgements
The authors wish to thank the National Council of Science and Technology (CONACyT) for the support scholarship granted to H. García-Núñez for her PhD studies; to the Secretariat of Public Education for funding the PIFI program to conduct this research project; to the Hispano-Luso Institute of Agricultural Sciences of the University of Salamanca, Spain, for its support to conduct the molecular studies and to the reviewers, especially the Associate Editor who patiently contributed to improve the structure of the document.
REFERENCES
Al-Mughrabi K. 2008. Biological Control of Phytophthora infestans of Potatoes using Trichoderma atroviride. Pest Technology. 2 (2): 104-108. http://dx.doi:10.3923/PPJ.2005.5.7 [ Links ]
Altieri A. 1999. The ecological role of biodiversity in agroecosystems. Agriculture, Ecosystems and Environment. 74: 19-31. http://dx.doi.org/10.1016/S0167-8809(99)00028-6 [ Links ]
Bailey A, Bae H, Strem M, Crozier J, Thomas S, Samuels G, Vinyard B and Holmes K. 2008. Antibiosis, mycoparasitism and colonization success for endophytic Trichoderma isolates with biological control potential in Theobroma cacao. Biocontrol 46(1):24-35. http://doi:10.1016/j.biocontrol.2008.01.003 [ Links ]
Barnett H and Hunter B. 1972. Ilustrated genera of imperfect fungi. EE. UU. Burgess Publ. Co. 241pp. http://dx.doi:10.2307/3757954 [ Links ]
Chaverri and Samuels G. 2003. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocraceae): species with green ascospores. Stud. Mycol.48:1-116. http://dx.doi:10.2307/3761915 [ Links ]
Chaverri P and Samuels G. 2002. Hypocrea lixii, the teleomorph of Trichoderma harzianum. Mycological Progress 1(3):283-286. http://dx.doi:0.1007/s11557-006-0025-8 [ Links ]
De la Cruz-Pérez, D., Ortiz-García, C. F., Bautista-Muñoz, C., Ramírez-Pool, J. A., Ávalos-Contreras, N., Cappello-García, S.,. (2015). Diversidad de Trichoderma en el agroecosistema cacao del estado de Tabasco, México. Revista Mexicana de Biodiversidad 86 (4), 947-961. http://dx.doi.org/10.1016/j.rmb.2015.07.012 [ Links ]
De los Santos-Villalobos, S., Guzmán-Ortiz, D. A., Gómez-Lim, M. A., Délano-Frier, J. P., de-Folter, S., Sánchez-García, P., and Peña-Cabriales, J. J. (2013). Potential use of Trichoderma asperellum (Samuels, Liechfeldt et Nirenberg) T8a as a biological control agent against anthracnose in mango (Mangifera indica L.). Biological Control, 64(1), 37-44. http://dx.doi.org/10.1016/j.biocontrol.2012.10.006 [ Links ]
Debebe Z, Amutha S, Dereje G, Mesfin T and Bekele K. 2011. Biocontrol activity of Trichoderma viride and Pseudomonas fluorescens against Phytophthora infestans under greenhouse conditions. Journal of Agricultural Technology. 7(6): 1589-1602. http://dx.doi:10.5423/PPJ.2004.20.1.075 [ Links ]
Druzhinina S. and Kubicek P. 2005. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? Journal of Zhejiang University Science. 6(2):100-112. http://dx.doi:10.1631/jzus.2005.B0100 [ Links ]
Erwin, D. C., and Ribeiro, O. K. 1996. Phytophthora Diseases Worldwide. The American Phytopathological Society, St. Paul, MN. http://dx.doi.org/10.1046/j.1365-3065-3059.1998.00179.x [ Links ]
FAO (Organización de las Naciones Unidas para la Agricultura y la Alimentación). 2010. Perspectivas Agrícolas OCDE-FAO. http://dx.doi:10.1787/agr_outlook-2010-2-es [ Links ]
García-Núñez H., Romero G, González E, Nava B y Martínez C. 2012. Isolation of Native Strains of Trichoderma spp. from horticultural soils of the Valley of Toluca, for potential biocontrol of Sclerotinia. Tropical and Subtropical Agroecosystems. 15:357-365. Disponible en linea:http://www.revista.ccba.uady.mx/ojs/index.php/TSA/article/ view/890. [ Links ]
Guigón-López C, Guerrero P, Vargas A, Carvajal M, Ávila Q, Bravo L, Ruocco L, Lanzuise S, Woo S. y Lorito M. 2010. Identificación Molecular de Cepas Nativas de Trichoderma spp., su Tasa de Crecimiento in vitro y Antagonismo contra Hongos Fitopatógenos. Revista Mexicana de Fitopatología. 28:87-96. Disponible en línea: http://www.redalyc.org/pdf/612/61218468002.pdf [ Links ]
Hall T. 2004. Biological sequence alignment editor. Ibis bioscience. Carlsbad Ca. Hammer O, Harper D. and Ryan P. 2001. PAST: Paleontological statistics software package for education and data analysis. Paleontology electronic 4(1):9. http://dx.doi:10.1002/9780470750711 [ Links ]
Harman G, Howell C, Viterbo A, Chet I, and Lorito M. 2004. Trichoderma species - opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2:43-56. Disponible en línea: www.ncbi.nlm.nih.gov/pubmed/15035008. http://dx.doi:10.1038/nrmicro797 [ Links ]
Harman G, Obregon M, Samuels G and Lorito M. 2010. Changing models for commercialization and implementation of biocontrol in the developed and developing world. Plant Disease 94: 928-939. http://dx.doi:10.1094/PDIS-94-80928 [ Links ]
Harris I. J.1986. Modified method for fungal slide culture. Journal of clinical microbiology. 24(3): 460-461. Disponible en línea:http://jcm.asm.org/content/24/3/460.full.pdf [ Links ]
Hermosa R, Grondona I, Iturriaga E, Díaz M, Castro C, Monte E y García A. 2000. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Applied Environmental Microbiology. 66: 1890-1898. http://dx.doi.10.1128/AEM.66.5.1890-1898.2000. [ Links ]
Jaklistch W, Komon M, Kubicek C, and Druzhinina I. 2005. Hypocrea voglmayrii sp. Nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 97 (6): 1365-1378. http://dx.doi:10.3852/mycologia.97.6.1365 [ Links ]
Kerroum F, Karkachi N, Jamal E y Kihal M. 2015. Antagonistic effect of Trichoderma harzianum against Phytophthora infestans in the North-west of Algeria. International Journal of Agronomy and Agricultural Research (IJAAR). 6 (4): 44-53. Disponible en línea: http://www.innspub.net [ Links ]
Kredics L, Hatvan L, Naeimi S, Körmöczi P, László M, Casaba V y Druzhinina I. 2014. Biodiversity of the Genus Hypocrea/Trichoderma in Different Habitats. En: Gupta G. et al. (eds.) Biotechnology and biology of Trichoderma. Elsevier, Amsterdam. pp. 3-24. http://dx.doi.org/10.1016/ B978-0-444-59576-8.00001-1 [ Links ]
Larralde C, Santiago M, Sifuentes R, Rodríguez L, Rodríguez P, Shirai K, and Narváez Z. 2008. Biocontrol potential and polyphasic characterization of novel native Trichoderma strains against Macrophomina phaseolina isolated from sorghum and common bean. Applied Microbiology and Biotechnology. 80: 167-177. http://dx.doi:10.1007/s00253-008-1532-0 [ Links ]
López C, Martínez A, Peñuelas G., Arteaga T, Ruiz A, Gómez T y Palmero D. 2013. Presencia de aislamientos homotálicos de Phytophthora infestans en cultivos de papa (Solanum tuberosum L.) tratados con metalaxil en el Valle Toluca, México. ITEA .109 (4):408-414. http://dx.doi.org/10.1094/PDIS-10-12-0962-PDN. [ Links ]
Lorito M, Woo S, Harman G and Monte E. 2010. Translational research on Trichoderma: from omics to the field. Annual Review of Phytopathology 48: 395-417. http//doi:10.1146/annurev-phyto-073009-114314. [ Links ]
Martínez, B., Solano T. 1994. Antagonismo de Trichoderma spp. frente a Alternaria solani (Ellis y Martin) Jones y Grout. Revista Protección Vegetal, 10: 221-22. Disponible en línea: http://www.sidalc.net/cgi [ Links ]
Matroudi S, Zamani M and Motallebi M. 2009. Antagonistic effects of three species of Trichoderma sp. on Sclerotinia sclerotiorum, the causal agent of canola stem rot. Egyptian Journal of Biology. 11: 37-44. Disponible en línea: http://ecology.nottingham.ac.uk/~plzfg/EBBSoc/ejb11/5-Matroudi_et_al_2009.pdf [ Links ]
Michel A, Otero S, Solano P, Ariza F, Barrios A y Rebolledo M. 2009. Biocontrol in vitro con Trichoderma spp., Fusarium subglutinans, (Wollenweb y Reinking) Nelson, Toussoun y Marasas y F. oxysporum Schlecht., agentes causales de la “Escoba de bruja” del Mango (Mangifera indica L.). Revista Mexicana de Fitopatología, 27. 18-26. Disponible en linea:http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S0185-33092009000100003 [ Links ]
Miller J and Johnson D. 2000. Competitive fitness of Phytophthora infestans isolates under semiarid field conditions. Phytopathology 90: 220-227. http://dx.doi.org/10.1094/PHYTO.2000.90.3.220 [ Links ]
Mohammad S, Mukesh S, Kumar V, Singh A, Sharma A, Sonika P, Rastogi S, Neelam P and Srivastava A. 2014. Phylogenetic diversity analysis of Trichoderma species based on internal transcribed spacer (ITS) marker. African Journal of Biotechnology 13(3):449-455. http://dxdx.doi.org/10.5897/ajb2013.13075 [ Links ]
Monte G. (2001) Understanding Trichoderma: between biotechnology and microbial ecology. International Microbiology. 4 (1): 1-4. http://dx.doi.10.1007/s101230100001 [ Links ]
Morrone J. 2001. Sistemática, biogeografía y evolución. México: UNAM. http://dx.doi:10.1086/421671 [ Links ]
Osorio-Concepción M., Casas F. S., Cortés P. C. 2013. Efecto de la limitación de fosfato sobre la conidiación de Trichoderma atroviride y mutantes ciegas a la luz. Revista Mexicana de Micología 37: 41-40. http://dx.doi:10.1016/j.riam.2011.11.002 [ Links ]
Osorio-Nila, M. A., Vázquez-García, L. M., Salgado-Siclán, M. L., González-Esquivel, C. E (2005). Efecto de dos enmiendas orgánicas y Trichoderma spp. para controlar Sclerotinia spp. en lechuga. Revista Chapingo Serie Horticultura. 11(3):203-208.http://dx.doi:10.5154/r.rchsh.2013.07.023 [ Links ]
PHC (Plant, Health Care Mex.) Disponible en www.agroquimicos-organicosplm.com/phc-t-22-1645-9 (Consulta, septiembre 2016). http://dx.doi.org/10.7748/phc.13.10.7.s7 [ Links ]
Sadfi Z, Hannachi I, Rouaissi M, Hajlaoui M, Rubio M, Monte E, Boudabous A. and Hermosa R. 2009. Biodiversity of Trichoderma strains in Tunisia. Canadian Journal of Microbiology. 55:154-162. http://dx.doi.10.1139/w08-101. [ Links ]
Samaniego-Gaxiola, J.A. 2008. Efecto del pH en la sobrevivencia de esclerocios de Phymatotrichopsis omnívora (Dugg.) Hennebert expuestos a Tilt y Trichoderma sp. Revista Mexicana de Fitopatología, 26: 32-39. http://dx.doi.org/10.7550/rmb.37737. [ Links ]
Samuels G. J., Adnan I., Marie C. B., Sophie De R., Orlando P. 2010. Trichoderma asperellum sensu lato consists of two cryptic species. Mycologia.102(4). 944-966. http://dx.doi:10.3852/09-243 [ Links ]
Schuster, A., Schmoll, M. (2010). Biology and biotechnology of Trichoderma. Applied Microbiology and Biotechnology, 87(3), 787-799. http://dx.doi:10.1007/s00253-0102632-1 [ Links ]
SIAP (Servicio de Información y Estadística Agroalimentaria y Pesquera). 2014. Anuario estadístico de la producción agrícola de los Estado Unidos Mexicanos. http://dx.doi:10.1787/agr_outlook-2011-es [ Links ]
Taghdi Y, Hermosa R, Domínguez S, Rubio M, Essalmani H, Nicolás C. and Monte E. 2015. Effectiveness of composts and Trichoderma strains in the control of Fusarium wilt of tomato. Phytopathology Mediterranea, 54(2), 232-240. http://dx.doi:10.14601/Phytopathol_Mediterr-15226 [ Links ]
Weindling R. 1932. Trichoderma lignorum as a parasite of other soil fungi. Phytopathology. 22: 837-845. http://dx.doi:org/10.1094/phyto-71-282 [ Links ]
Woo S, Scala F, Ruocco M, and Lorito M. 2006. The molecular biology of the interactions between Trichoderma spp., phytopathogenic fungi, and plants. Phytopathology 96:181-185. http://dx.doi.10.1094/Phyto-96-0181. [ Links ]
Received: May 11, 2016; Accepted: October 13, 2016











 text in
text in