INTRODUCTION
Tumor inhibitor genes are considered to exert a critical role in cancer development1. Genetic changes in these genes contribute to the loss of their functions, potentially resulting in unlimited cell proliferation, elevated cell invasion and migration abilities, cell resistance to a hypoxic environment, and cell metabolic remodeling2. Furthermore, some microRNAs (miRNAs) can inhibit the expression of tumor suppressor genes post-transcriptionally, promoting tumor progress.
MiRNAs, usually composed of 18-22 nucleotides, can modulate the expression of target genes through binding with the complementary sequences on the 3’untranslated region (3’-UTR) within mRNA, thereby inhibiting mRNA translation or inducing mRNA to become unstable3. As described above, miRNAs could affect malignant phenotypes of various cancers4-7; therefore, they can be considered potential biomarkers for predicting tumor occurrence, progression, and patient’s prognosis.
Recently, some scholars found that microRNA-421 (miR-421) is implicated in cell growth and exerts an essential effect on cancer occurrence and progression8. Studies reported a certain mediating effect on hepatocellular carcinoma (HCC) progression by miR-421. MiR-421 can be used as a marker of liver cancer. For example, Xie et al.9 analyzed the competitive endogenous (ceRNA) network constructed by HCC samples and found that the expression of miR-421 is associated with the prognosis of HCC patients and it may be one of the candidates for HCC markers. In another paper, they also found that miR-421 is associated with HCC10. In the study of cancer progression, miR-421 is involved in HCC progression as a member of the ceRNA network. For example, Li et al.11 found that the circITCH/miR-421/BTG1 axis inhibits HCC development and overexpression of circITCH or BTG1 can eliminate the malignant phenotype of HCC cells caused by miR-421 overexpression. Zhang et al.12 revealed that the hsa_circ_0003528/miR-421 axis can stimulate the epithelial-mesenchymal transition (EMT) process of HCC cells. In addition, miR-421 can foster HCC cell proliferation and migration through down-regulating the human farnesoid X receptor13. Many studies have shown that miR-421 may be an oncogene in HCC, but its potential mechanism in HCC remains to be fully elucidated.
4-aminobutyrate aminotransferase (ABAT) encodes the protein that can metabolize gamma aminobutyric acid into succinic semialdehyde14. ABAT plays an important role in tumorigenesis and immunity. Studies have found that low expression of ABAT is associated with poor prognosis of HCC and can be used as an independent prognostic factor for HCC. Its expression is related to glycolysis-related genes and immune cell infiltration in HCC15. This suggests that ABAT may also play an important role in the occurrence and immunity of HCC. However, no conclusion is definite about its biological role in HCC. This study identified the targeting relationship between ABAT and miR-421, and then investigated the biological roles of miR-421/ABAT regulatory axis in HCC cells and proposed a novel modulatory mechanism for HCC in vitro.
MATERIALS AND METHODS
Bioinformatics methods
Expression data of mature miRNAs (normal: 50, tumor: 375) and mRNA (normal: 50, tumor: 374) in were obtained from The Cancer Genome Atlas-Liver HCC (TCGA-LIHC) database. Differential expression and survival significance of miR-421 and ABAT were analyzed based on the downloaded data. The "edgeR" package was used to conduct differential analysis of the mRNAs (|logFC| >1.5, padj < 0.01) to get differentially expressed mRNAs (DEmRNAs). The TargetScan, mirDIP, and starBase databases were utilized to predict potential target mRNAs of miR-421. In addition, the predicted mRNAs were overlapped with the downregulated DEmRNAs in TCGA, and the most relevant mRNA was selected for follow-up research.
Cell culture
Normal human hepatic cell line HL-7702 (BNCC100012) and human HCC cell lines SMMC-7721 (BNCC338089), HepG2 (BNCC338070), Huh-7 (BNCC337690), and HLE (BNCC100966) were accessed from BeNa Culture Collection (BNCC, China). HL-7702 cell line was cultured in CM2-1 culture medium with 5% CO2 at 37°C. CM2-1 culture medium: 90% RPMI medium-1640 + 10% FBS. SMMC-7721, HepG2, and Huh-7 cells were maintained in CM1-1 culture medium with 5% CO2 at 37°C. CM1-1 culture medium: 90% high glucose DMEM + 10% FBS. HLE cell line was cultured in 90% RPMI-1640 + 10% FBS with 5% CO2 and 95% air at 37°C. All media were purchased from BNCC (China).
Cell transfection
miR-421 inhibitor (miR-inhibitor) and negative control inhibitor (NC-inhibitor), si-ABAT, and control si-NC were bought from RiboBio (China). Following the protocol of Lipofectamine 2000 Kit (Invitrogen, USA), synthesized reagents were transfected into HCC cells.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cells with TRIzol reagent (Thermo Fisher Scientific, Inc., USA), and miRNA and mRNA were, respectively, reversely transcribed into cDNA with miScript reverse transcription Kit (Bio-Rad Laboratory, USA). SYBR-Green method (Qiagen, Inc., Germany) was used to detect relative levels of target genes on the Bio-Rad CFX96 qRT-PCR system. PCR was performed at 95°C for 15 min, and then for 40 cycles of 94°C for 15 s, 55°C for 30 s, and 63°C for 30 s. GAPDH and U6 were utilized as internal references. Levels of miR-421 and ABAT mRNA were analyzed by the 2-ΔΔCT method. The assay was fulfilled in triplicate and conducted independently. Primer sequences are listed in Table 1.
Table 1. Primer sequences for qRT-PCR
| Name | Forward primer (5’->3’) | Reverse primer (5’->3’) |
|---|---|---|
| miR-421 | GTCGCGCGGGUUAAUGCCTC | GGACATUAGUUGUCUGUAAATAG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| ABAT | AAGAGAGCCGAGGCAATTACC | GCTCGCATTTTGAGGCTGTTG |
| GAPDH | GACCTGACCTGCCGTCTA | AGGAGTGGGTGTCGCTGT |
qRT-PCR: quantitative reverse transcription polymerase chain reaction.
Cell proliferation assay
CCK-8 analysis method (Beyotime Biotechnology, China) was commended. 2 × 103 cells were inoculated into each well of a 96-well plate. At 0, 24, 48, and 72 h, 10 μL of CCK-8 solution was dropped into each well, respectively. After maintaining at 37°C for 1.5 h, absorbance at 450 nm was assayed by the Spectro Max 250 spectrophotometer (Molecular Devices, USA). The assay was carried out in triplicate and conducted independently.
Cell invasion assay
Cells were digested with trypsin and resuspended overnight in serum-free DMEM. To detect the invasion level, 3 × 104 cells were plated into the upper insert coated with Matrigel and cultured in 600 ul DMEM. The medium with 10% FBS as a chemical attractant was supplemented to the lower chamber, with six repeated wells in each group. Twenty-four h later, the remained cells were wiped with cotton swabs. Cells beneath the lower surface were subjected to fixing with 4% paraformaldehyde for 30 min, staining with 0.1% crystal violet for 10 min, washing with double distilled water, and then drying naturally. Then, cells were observed using a microscope and photos of random fields were taken within 5 min. The number of penetrating cells was counted.
Wound healing assay
All instruments were sterilized and 2 × 105 cells were planted in a 6-well plate. When the cell coverage rate was 80%, the tip of a 200 μl pipette was utilized to scrape the cell monolayer through the well center, and then cells were rinsed with phosphate-buffered saline 3 times. Scraped cells were discarded and serum-free medium was then supplemented on the plate. The cells were put into an incubator with routine conditions for culture. Images were taken at 0 and 24 h. The wound healing rate was computed as per the formula: Wound healing rate = (W0 − W24) / W0 × 100% (W0 was the wound width at 0 h; W24 was the wound width at 24 h).
Flow cytometry
Flow cytometry was used to detect cell cycle and apoptosis. Details are shown in the supplementary material.
Dual-luciferase reporter gene assay
A dual luciferase reporter gene detection kit (Promega, USA) was applied to measure luciferase activity. Details are shown in the supplementary.
Western blot
BCA kit (Thermo Fisher Scientific, USA) was recommended to assay the concentration of isolated proteins from cells. The information on antibodies is exhibited in supplementary table 1.
Lactic acid and NADP/NADPH quantification analysis
For lactic acid measurement, Lactate assay kit (Sigma-Aldrich, USA) was employed. Details are shown in the supplementary.
Patients
The clinical information of liver cancer patients was downloaded from TCGA database. Patients were included in the analysis according to the following criteria: imaging techniques and post-operative pathology confirmed HCC; absence of coeval tumors; without signs of infection; and completed clinical and follow-up data.
Exclusion criteria of patients: patients without complete clinical information. Only patients without survival status and survival time were excluded when drawing the survival curve. According to the median value of miR-421 expression, the samples were divided into high- and low-expression groups. The clinicopathological characteristics of HCC patients are shown in table 2.
Table 2. Clinicopathological characteristics of HCC patients
| Characteristic | Low expression hsa-miR-421 (n = 183) | High expression hsa-miR-421 (n = 182) | p-value |
|---|---|---|---|
| Age (years) | |||
| ≤ 65 | 122 (66.7%) |
110 (60.4%) |
0.26 |
| > 65 | 61 (33.3%) |
72 (39.6%) |
|
| Gender | |||
| Female | 60 (32.8%) |
55 (30.2%) |
0.678 |
| Male | 123 (67.2%) |
127 (69.8%) |
|
| T | |||
| T1 | 102 (55.7%) |
80 (44.0%) |
0.162 |
| T2 | 39 (21.3%) |
51 (28.0%) |
|
| T3 | 36 (19.7%) |
44 (24.2%) |
|
| T4 | 6 (3.3%) |
7 (3.8%) |
|
| N | |||
| N0 | 118 (64.5%) |
135 (74.2%) |
0.0538 |
| N1 | 1 (0.5%) |
3 (1.6%) |
|
| NX | 64 (35.0%) |
44 (24.2%) |
|
| M | |||
| M0 | 128 (69.9%) |
138 (75.8%) |
0.329 |
| M1 | 3 (1.6%) |
1 (0.5%) |
|
| MX | 52 (28.4%) |
43 (23.6%) |
|
| G | |||
| G1 | 35 (19.1%) |
19 (10.4%) |
0.0108 |
| G2 | 94 (51.4%) |
82 (45.1%) |
|
| G3 | 48 (26.2%) |
74 (40.7%) |
|
| G4 | 6 (3.3%) |
7 (3.8%) |
HCC: hepatocellular carcinoma.
Statistical analysis
R software (version 4.2.1, Lucent Technologies, USA), GraphPad Prism 7.04 (GraphPad Inc., USA), and Flow-Jo V7 software (TreeStar, USA) were utilized to process data. Pearson correlation coefficient (PCC) was utilized to assay the correlation between ABAT and miR-421. Log-rank was utilized to compare differences in the overall survival time of patients. Kruskal–Wallis was used to analyze differences in data between multiple groups.. Welch’s t-test and Wilcoxon rank-sum test were utilized to test the difference between groups. All experiments were repeated 3 times. p < 0.05 was thought to be statistically significant.
RESULTS
High expression level of miR-421 in HCC tissue and cells
The previous studies have shown that miR-421 is significantly overexpressed in HCC cells, and its upregulation promotes HCC cell viability and colony formation while reducing cell apoptosis11,12. Based on the above result, we are assuming that miR-421 may serve as an onco-miRNA in HCC. First, we downloaded the expression data and clinical information of miR-421 in HCC patients from TCGA database. According to the median value of miR-421 expression, HCC patients were divided into high and low expression groups of miR-421. Among them, there were 185 and 186 patients in the high- and low-expression groups of miR-421 including survival time and survival status, respectively. The clinical baseline characteristics of these patients are shown in table 2. Based on these patients, we used Kaplan–Meier (K-M) survival curve to explore the relationship between the expression of miR-421 and the overall survival rate of patients. Survival analysis has been widely applicated to assess disease progression and treatment efficiency in clinical practice16. The survival analysis indicated that patients’ overall survival time in the high miR-421 level group was shorter relative to that of patients in the low miR-421 level group (Figure 1A, p < 0.001, 95% CI = 1.41-2.815). Then, when we analyzed the expression status of miR-421, we found that it was noticeably increased in HCC tissues (Figure 1B, p < 0.05). Analysis of the relationship between miR-421 and TNM stage or grade of HCC revealed that the expression of miR-421 was positively correlated with high grade HCC (p < 0.05), but had no relationship with TNM stage (p > 0.05) (Figure 1C). Based on the above bioinformatics analysis, we further validated miR-421 at the cellular level. We manifested that miR-421 was notably upregulated in cancer cells, with the most significant upregulation in SMMC-7721 and HepG2 cells, as detected by qRT-PCR (Figure 1D, p < 0.05). Therefore, HepG2 and SMMC-7721 cells were used for follow-up assays. Taking the above results together, the high level of miR-421 was demonstrated in HCC tissues and cells.
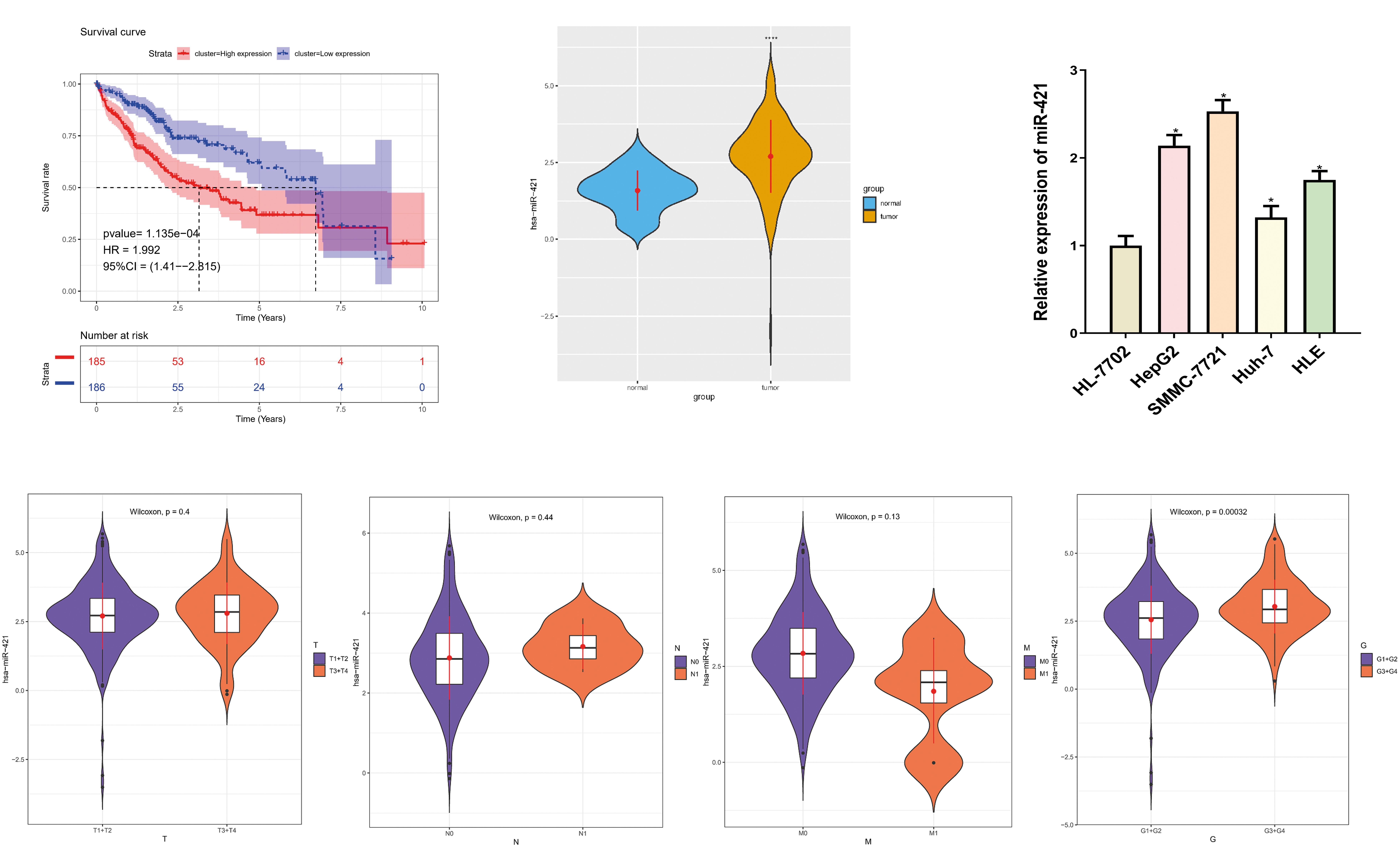
Note: *Indicates, p < 0.05.
Figure 1. MiR-421 is increased in HCC. (A) Relationship between miR-421 level and survival rate of patients with HCC (red represents the high expression of miR-421, blue represents the low expression of miR-421); (B) Expression of miR-421 in HCC tissues in TCGA; (C) The relationship between miR-421 expression and clinical stages; (D) miR-421 level in HCC cell lines assayed through quantitative reverse transcription polymerase chain reaction.
Suppressing miR-421 expression attenuates the proliferation, migration, and invasion of hepatocellular carcinoma cells
HCC cells were classified into two groups to assay the impact of miR-421 on HCC cell biological functions, including miR-inhibitor and NC-inhibitor groups, and the transfection efficiencies were assayed through qRT-PCR. MiR-421 was notably less expressed in the miR-inhibitor group compared to that in the NC-inhibitor group (Figure 2A, p < 0.05). Then, the CCK-8 result manifested that cell proliferation of the miR-inhibitor group was remarkably downregulated (Figure 2B, p < 0.05). Next, Transwell assay was utilized to assay cell invasive property. Similarly, the invasive potential of cells was significantly downregulated after miR-421 expression was suppressed (Figure 2C, p < 0.05). Finally, wound healing assay was done to assay the cell migratory property of the two groups. Consistent with the previous experimental results, the migratory ability of cells was also significantly downregulated after silencing miR-421 (Figure 2D, p < 0.05). The influence of miR-421 on the HCC cell cycle was also analyzed. Knockdown of miR-421 markedly elevated the proportion of cells in G0/G1, remarkably decreased the proportion of cells in the S phase (Figure 2E, p < 0.05), and notably increased the apoptosis rate (Figure 2F, p < 0.05). Among proteins associated with the EMT process, E-cadherin protein expression was noticeably increased, and Vimentin protein expression was substantially decreased (Figure 2G, p < 0.05). Based on these results, the miR-421 level was positively related to the malignant progression of HCC cells.
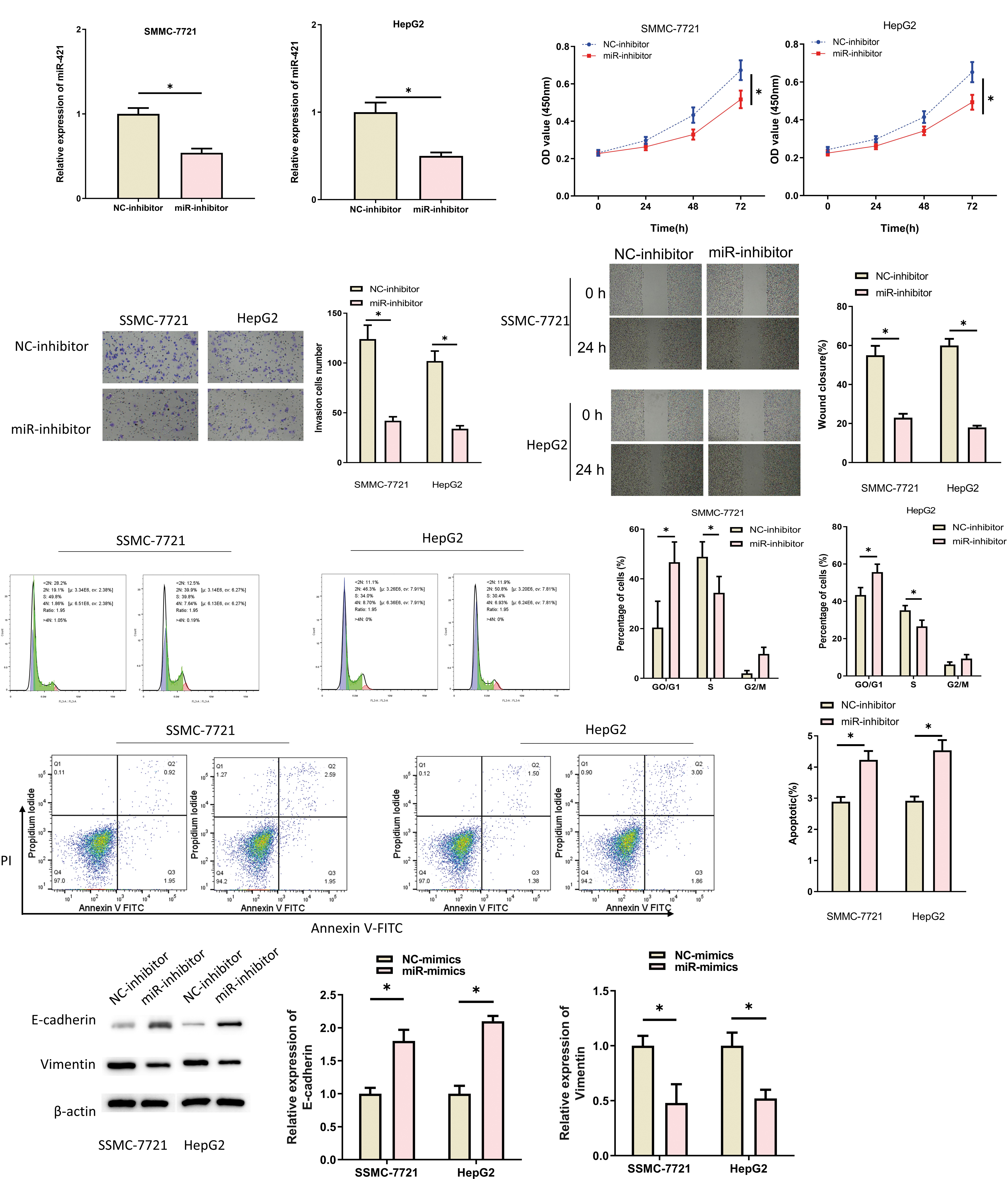
Note: *Indicates, p < 0.05.
Figure 2. Suppressing miR-421 expression attenuates the malignant process of HCC cell. A: MiR-421 level in miR-inhibitor group and NC inhibitor group; B: Proliferation of HCC cells in two groups; C: Invasive potential of HCC cells in two groups; D: Migratory potential of HCC cells in two groups. E: Cell cycle of HCC cells in two groups; F: The apoptotic rate of HCC cells in two groups; G: Expression of EMT-related proteins of HCC cells in two groups.
miR-421 downregulates ABAT expression in hepatocellular carcinoma cells
To verify the downstream mechanisms of miR-421, TargetScan, mirDIP, and starBase databases were employed for identifying possible targets of miR-421, and then results were overlapped with downregulated DE mRNAs in TCGA (Figure 3A). Through correlation analysis, ABAT with the highest association with miR-421 was selected as the target (Figure 3B). The bioinformatics analysis disclosed that ABAT level was substantially low in HCC tissues than in normal ones (Figure 3C, p < 0.05). We reasoned that ABAT expression may correlate with the TNM stage and grade of HCC patients. The results pointed out that ABAT expression was positively correlated with advanced T stage and high grade (p < 0.05), but had no relationship with N and M stages (p > 0.05) (Figure 3D). To further verify the bioinformatics prediction, we verified the targeting relationship of miR-421 and ABAT using dual-luciferase assay. The result manifested that the luciferase activity of cell lines co-transfected with miR-inhibitor and WT-ABAT 3’-UTR was remarkably increased, but that of cell lines cotransfected with MUT-ABAT 3’-UTR had no change (Figure 3E-F, p < 0.05). Next, ABAT mRNA and protein levels in normal human hepatic cells and HCC cells were assayed through qRT-PCR and western blot, respectively. ABAT level was decreased in HCC cells, and cell line HepG2 was selected for the following experiments (Figure 3G-H, p < 0.05). In addition, K-M analysis on ABAT expression level was introduced, whose results presented that the patients with high ABAT expression shared relatively better survival statuses (Figure 3I). Then, qRT-PCR (Figure 3J, p < 0.05) and western blot (Figure 3K, p < 0.05) were utilized to assess ABAT mRNA and protein levels when miR-421 was knocked down, showing that ABAT was conspicuously upregulated at mRNA and protein levels. These findings manifested that ABAT was targeted by miR-421 in HCC cells, and miR-421 could downregulate ABAT expression.
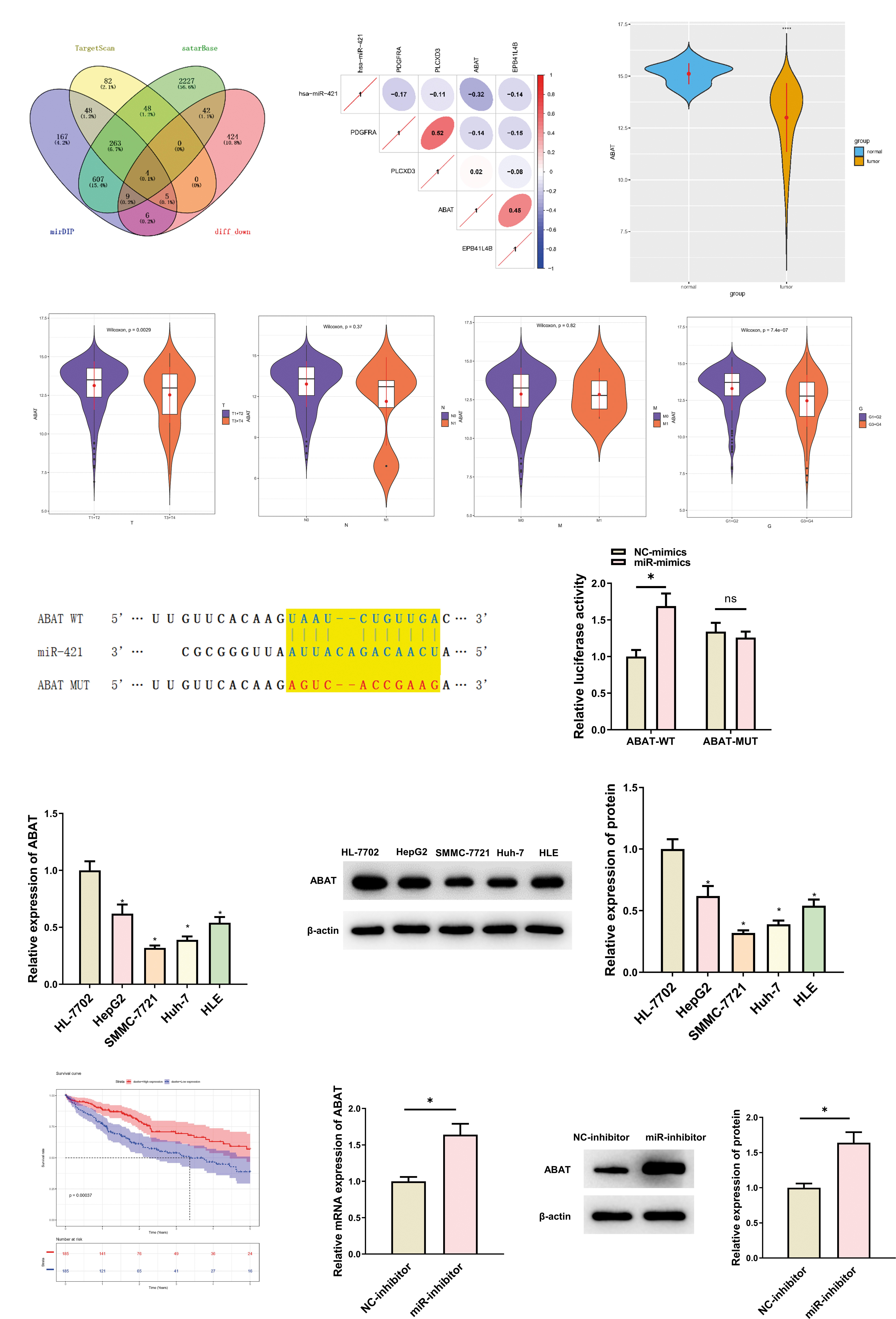
Note: *Indicates, p < 0.05.
Figure 3. The targeting relationship of miR-421 and ABAT. A: The intersection of predicted target genes of miR-421 and the decreased DEmRNAs in TCGA; B: Pearson correlation analysis of miR-421 and ABAT; C: Relative expression level of ABAT in TCGA; D: Relationship of ABAT expression with HCC clinical stages; E: The putative targeted binding sites of miR-421 and ABAT; F: The luciferase activity of ABAT-WT group and ABAT-MUT group; G-H: mRNA and protein levels of ABAT in normal human hepatic cell line and HCC cell lines; I: K-M analysis on ABAT level for HCC patients (red represents the high expression of ABAT, blue represents the low expression of ABAT); J-K: mRNA and protein levels of ABAT in the NC-inhibitor group and miR-inhibitor group.
miR-421/ABAT axis affects HCC cell proliferation, migration, and invasion
To prove whether miR-421 could foster the malignant phenotype of HCC cells through targeting ABAT, HCC cells were either transfected with si-ABAT alone or cotransfected with si-ABAT and miR-421 inhibitor. qRT-PCR was utilized to assess ABAT level to ensure cotransfection efficiencies. ABAT was substantially downregulated in cells in which ABAT expression was silenced alone, but transfection of miR-inhibitor reversed the suppressing impact of si-ABAT in HCC cells (Figure 4A, p < 0.05). CCK-8, wound healing, and Transwell assays were utilized to evaluate cell proliferation, migration, and invasion in NC-inhibitor + si-NC, NC-inhibitor + si-ABAT, and miR-inhibitor + si-ABAT groups. Compared with the NC-inhibitor + si-NC group, proliferative, migratory, and invasive properties of HCC cells with only silencing ABAT expression were strengthened, while additionally transfecting miR-inhibitor could reverse the strengthened effects (Figure 4B-D, p < 0.05). Flow cytometry presented that silencing of ABAT decreased the proportion of G0/G1 phase cells, increased the proportion of S phase cells, and reduced apoptosis rate, but these effects were repressed by concomitant inhibition of miR-421 expression (Figure 4E and F, p < 0.05). EMT-related protein expressions exhibited the same trend. Silencing of ABAT increased E-cadherin protein expression and decreased Vimentin protein expression, while concomitant inhibition of miR-421 repressed these effects (Figure 4G, p < 0.05). Lactic acid concentration and NADPH/NADP homeostasis are two prominent features of the tumor microenvironment (TME), which are closely related to tumor metabolism. The metabolic state of the tumor can affect the occurrence and progression of tumor cells17,18. Therefore, the metabolic status of tumor cells was evaluated. Lactate production and NADPH/NADP+ were measured, and their results presented that suppressing ABAT expression could promote HCC cell metabolic remodeling, while miR-inhibitor reversed the promotion (Figure 4H, p < 0.05). Hence, the increase of miR-421 level facilitated cell proliferation, migration, invasion, and EMT process, inhibited apoptosis, and affected the metabolic remodeling of HCC cells, but decreased ABAT expression.
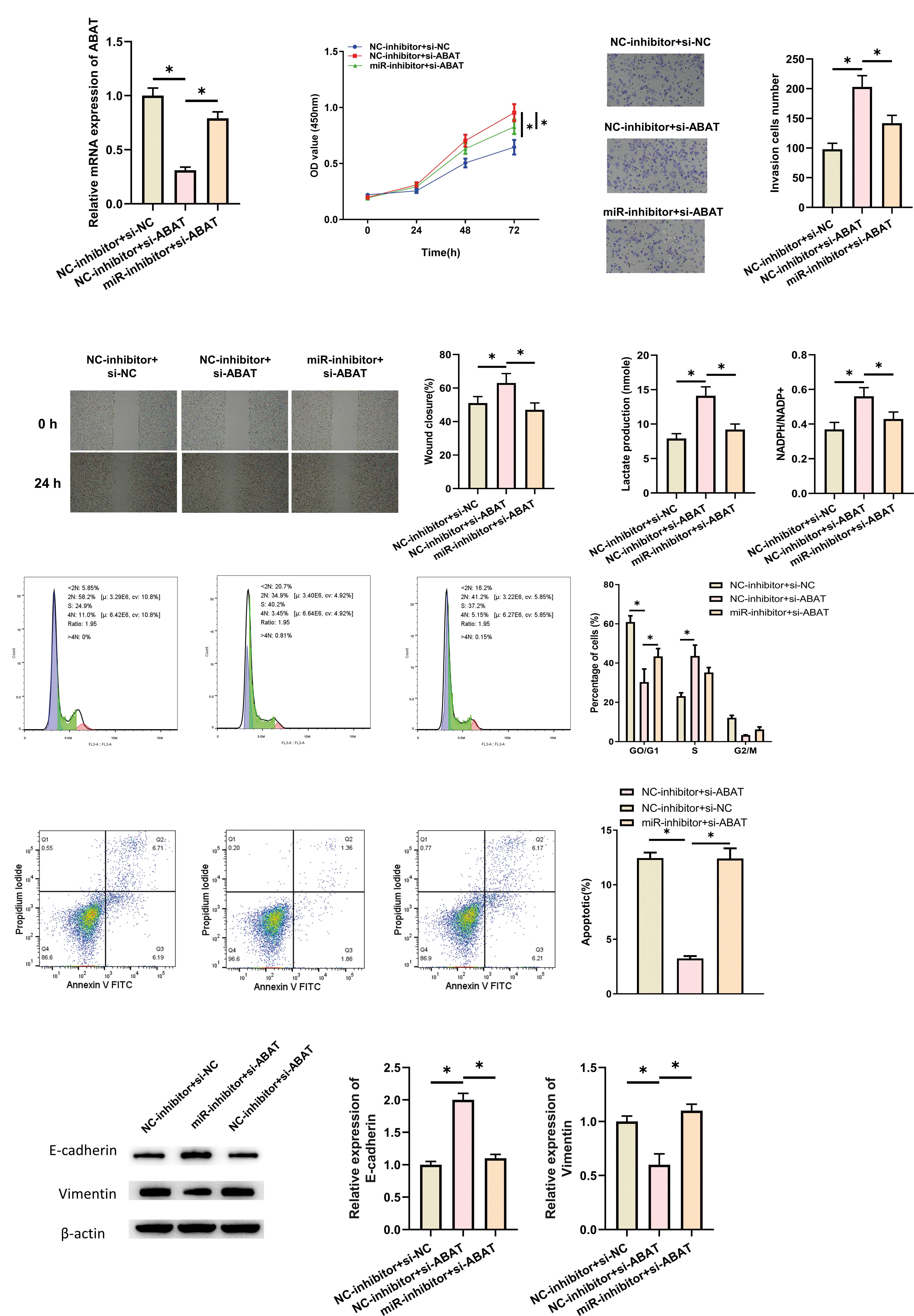
Note: *Indicates, p < 0.05.
Figure 4. miR-421 targets ABAT, fosters phenotype progression, and affects the metabolic remodeling of HCC cells. A: ABAT level in NC-inhibitor + si-NC, NC-inhibitor + si-ABAT, and miR-inhibitor + si-ABAT groups assayed through quantitative reverse transcription polymerase chain reaction; B: HCC cell proliferative property assayed through CCK-8; C: HCC cell invasive property assayed through Transwell assay; D: HCC cell migratory property detected by wound healing assay; E: Cell cycle as tested by flow cytometry; F: Cell apoptosis as tested by flow cytometry; G: EMT-related protein expression as assayed by western blot; H: The lactate production and NADPH/NADP+ were analyzed at 450 nm.
DISCUSSION
It is well known that the mortality of HCC is extremely high. Despite progress in screening and treatment, HCC incidence remains on the rise19. China is the most severely influenced country, with an age-standardized incidence rate of 22.3/100,000 (male: 33.7/100,000, female: 10.9/100,000). HCC is the second leading cause of death of cancer in China, with a mortality of 21.4/100,000 (male: 32.3/100,000, female: 10.7/100,000), accounting for 17.4% of total cancer deaths20. The current treatment for HCC is very limited and the survival rate after diagnosis remains very low. Hence, it is urgent to deeply know the molecular mechanisms of HCC cancer to develop therapeutic strategies.
At present, the clinical research on miR-421 is involved in a wide range of studies and reports that miR-421 is noticeably increased in a number of cancer species, such as esophageal adenocarcinoma21, NSCLC22,23, and breast cancer24. Besides, we used the K-M curve to explore the relationship between the expression of miR-421 and the survival time of patients. High miR-421 expression in tumor tissue is associated with poor overall survival, displaying the possible role of miR-421 as a target for cancer therapy25. Therefore, miR-421 was selected here to clarify its role in HCC. First of all, we displayed that miR-421 was noticeably increased in tumor tissue in TCGA-LIHC dataset. Its expression was correlated with the clinical stage and its high level indicated a poor prognosis. To confirm the results of bioinformatics analysis, normal human hepatic cells and HCC cells were selected for research. We disclosed that miR-421 was markedly elevated in HCC cells as compared to normal hepatic cells, which was congruous with the result of bioinformatics analysis. Hui et al.26 illustrated that miR-421 was upregulated in HCC, which is in line with the findings of this study. In their study, regulatory role of miR-421 in the HCC malignant progression was not investigated, but we deeply explored influence of miR-421 on proliferation, migration, invasion, cell cycle, and apoptosis of HCC cells.
The previous studies unveiled that miRNA takes a vital part in cancer metastasis27-29. Our study found that inhibition of miR-421 level fostered phenotype progression of HCC cells and influenced the EMT process of HCC. To find the regulatory mechanism of miR-421 in HCC, bioinformatics methods were first applied to dig target genes of miR-421, and a regulatory axis of miR-421/ABAT was identified. As for ABAT, a study reported that the coexpression of ABAT and lncRNA is the regulator of myelodysplastic syndrome30. Reduced expression of ABAT and STC2 is a marker of ER-positive inflammatory advanced breast cancer and endocrine therapy resistance31. Song et al.32 performed clustering analysis for differential genes and key modules associated with clinical features of HCC through WGCNA. By network topology analysis, new risk genes were found from the key module and biologically validated, and it was uncovered that ABAT is closely relevant to tumor activity32. In our study, ABAT levels in HCC cells and normal hepatic cells were evaluated, and we displayed that ABAT in HCC cells was remarkably decreased. Cell functional experiments demonstrated that proliferative, migratory, and invasive abilities of HCC cells were remarkably strengthened, and apoptosis was remarkably inhibited while suppressing ABAT expression alone in HCC cells, yet these effects were recovered through inhibiting miR-421 expression. In all, it was verified that miR-421 could affect the malignant phenotype of HCC cells by regulating ABAT. Obviously, based on previous studies, this study proposed a new regulatory axis, miR-421/ABAT axis, and demonstrated its influence on HCC malignant progression. Nevertheless, our study is limited to in-vitro experiments. We do not know through which signaling pathways the miR-421/ABAT axis affects the cellular phenotype, and whether the miR-421/ABAT axis affects other characteristics, such as angiogenesis and the immune microenvironment; in addition, in vivo, whether the miR-421/ABAT axis also affects HCC cell growth and tumor metastasis, etc. These need to be investigated in cells and animals in combination with literature and bioinformatics analysis in future studies. Finally, we still lack the verification of clinical samples, which require us to collect more HCC clinical samples to verify and analyze our results. Despite multiple studies on the mechanism of miR-421 in HCC, our study further highlighted the important role of miR-421 in HCC and enriched the knowledge about mechanism of miR-421 in HCC.
Moreover, recurrence or metastasis is implicated in unfavorable prognosis in patients with HCC33. In this study, by analyzing TCGA database, the results of survival analysis on miR-421 illustrated that the overall survival time of patients in the miR-421 high expression group was substantially shorter than in the miR-421 low expression group. This result indicated that miR-421 high expression markedly predicted poor prognosis of HCC patients, indicating that miR-421 is a possible biomarker or clinical therapeutic target, which offers a fresh direction for diagnosis and targeted therapy of HCC patients.
In conclusion, the increased expression of miR-421 participated in HCC occurrence and progression by targeting and inhibiting ABAT expression, thus promoting the proliferation, migration, and invasion of HCC. Therefore, miR-421 and ABAT may be novel research directions for HCC therapy.











 text new page (beta)
text new page (beta)


