INTRODUCTION
Lung cancer, a leading cause of cancer-related deaths, has overtaken breast cancer as the biggest killer in developed regions1. Non-small cell lung cancer (NSCLC) made up approximately 85% of lung cancer cases, where lung adenocarcinoma (LUAD) is the major histological subtype2. Apart from its high incidence rate, for most patients, LUAD is often diagnosed when radical surgical resection is no longer possible. Although targeted molecular therapy has become a crucial anti-cancer therapy in fighting against advanced NSCLC, cancer drug resistance, metastasis, and recurrence kept poor long-term survival from improving2. Some light on the molecular mechanism of pathology has yielded tangible benefits in clinics by identifying the effective and therapeutic biomarkers in LUAD.
Tumor tissue is characterized by hypoxia and hypotrophy during tumor development3. Reportedly, under such conditions, activation of cancer-cell secreted angiogenic factors promotes massive neovascularization and provides the oxygen and nutrients needed for tumor growth4,5. The angiogenic processes include breakdown of basal lamina and extracellular matrix, migration and proliferation of endothelial cells (ECs), sprouting and branching of new vessels, and vessel maturation6. Angiogenesis is considered to be the main mechanism of tumor vascularization throughout the development, growth, invasion, migration, and distant metastasis of lung cancer7,8. Cong et al.2 elucidated that LINC00665 induced LUAD tumor-associated angiogenesis by interacting with YB-1 and activating the YB-1-angpt4/ANGPTL3/VEGFA axis. Lu et al.9 showed that anlotinib inhibited angiogenesis by suppressing CCL2 in NCI-H1975-derived xenograft models, whereas reduced CCL2 expression was associated with overall survival and progression-free survival benefit in patients with advanced NSCLC. These findings suggest that tumor angiogenesis affects the malignant progression of LUAD.
CTHRC1 is a chondrocyte-specific secreted glycoprotein with hormone-like characteristics10 that is involved in tissue remodeling processes affecting vascular and bone formation. CTHCR1 expressed in injured arteries is involved in remodeling of the adventitia of injured vessels by facilitating migration of fibroblasts and smooth muscle cells, and inhibiting collagen synthesis11. High CTHRC1 expression found in various cancers such as breast cancer, pancreatic cancer, oral squamous cell carcinoma, gastric cancer, and hepatocellular carcinoma12-16 is suggested to bear a tight relationship with the development of human cancers. The promoting role of CTHRC1 in tumor invasion and metastasis has also come to light in the previous studies. For example, in colorectal cancer metastasis, CTHRC1 is an intrinsic marker suggested by Zhang et al.17. They found that CTHRC1 could remodel infiltrating macrophages and promote colorectal liver metastasis through the TGF-β signaling pathway. Li et al.18 showed that, by altering recombinant CTHRC1 protein levels, endometrial tumor invasion and migration are promoted through enhancing macrophage recruitment in vitro.
In addition, CTHRC1 was demonstrated to be involved in regulating tumor angiogenesis. Lee et al.10 discovered that, as a potent endothelial activator, CTHRC1 induces the Ang-2/Tie2 axis to mediate recruitment of Tie2-expressing monocytes (TEMs) and promote angiogenesis in pancreatic cancer during tumorigenesis. However, the underlying mechanism of how high CTHRC1 expression affects angiogenesis in LUAD is poorly studied. Therefore, this study attempted to carry out an in-depth investigation of the molecular mechanism of CTHRC1 promoting angiogenesis in LUAD.
Homeobox B9 (HOXB9) belongs to the Homeobox (HOX) genes family and is the ninth paralog in the HOX-B cluster, implicated in the growth and progression of multiple cancers19. HOXB9 promotes the growth of LUAD through the AMPK-HOXB9-KRAS axis20. In pancreatic cancer, HOXB9 blocks cell cycle progression through the DNMT1/RBL2/c-Myc axis, thereby inhibiting the proliferation of cancer cells21. HOXB9 is overexpressed in colorectal cancer, which promotes proliferation and migration of cancer cells, and reduces the survival rate of colorectal cancer patients22. Nevertheless, the upstream regulatory gene of HOXB9 in LUAD remains unclear, and the role of the CTHRC1/HOXB9 axis in LUAD has not yet been studied.
The purpose of this study is to explore the biological function and mechanism of CTHRC1 in LUAD, so as to shed new light on the molecular mechanisms of LUAD progression.
MATERIALS AND METHODS
Bioinformatics analysis
mRNA expression data of LUAD (normal (N): 59, tumor (T): 539) were obtained from TCGA (https://portal.gdc.cancer.gov/). Subsequent differential analysis of mRNAs between N and T groups (| logFC | >2, padj < 0.05) using the edgeR package yielded 2516 DEmRNAs, of which 1996 were up-regulated and 520 down-regulated. The target genes were determined with the reference provided by the previous studies. GSEA software was used to perform target genes pathway enrichment analysis. Potential transcription factors upstream of target genes were predicted using KnockTF database (http://www.licpathway.net/KnockTF/index.html). The correlation between target genes and upstream transcription factors was analyzed by Pearson. Binding sites between target genes and upstream transcription factors were predicted by the online database JASPAR (http://jaspar.genereg.net/).
Cell culture and transfection
The human LUAD cell lines A549 (BNCC337696), NCI-H1975 (BNCC340345), PC-9 (BNCC340767), and human umbilical vein EC line (HUVECs) (BNCC342247) were procured from BeNa Culture Collection (BNCC, China). Normal lung epithelial cell lines BEAS-2B (CRL-9609) and 293T (CRL-3216) were bought from American Type Culture Collection (ATCC, USA). 293T cell line was used in dual luciferase reporter experiment. NCI-H1975 cell line was cultured in RPM1 1640 medium with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA). BEAS-2B cell line was cultured in BEGM media (Lonza, Walkersville, USA). A549 cell line was cultured in Kaighn's Modification of Ham's F-12 Medium (Thermo Fisher, USA). PC-9, 293T, and HUVECs cell lines were cultured in DMEM-H medium (Gibco, USA). The culture conditions of all cell lines were 37 Cº with 5% CO223.
Sh-NC/sh-CTHRC1 and sh-NC/sh-HOXB9, pGL3-Basic were used to construct oe-NC/oe-CTHRC1. All the plasmids were bought from RiboBio, China. The above vectors were transfected into LUAD cell lines using Lipofectamine 2000 kit (Thermo Fisher Scientific, USA) and cultured for 24 h before following experiments.
Chemical reagents
Orlistat, an inhibitor of FA (Ro18-0647)24 was purchased from Selleckchem, USA. Orlistat substance was prepared in dimethyl sulfoxide (DMSO). The control was treated with the same amount of DMSO.
Quantitative real time-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from different cells using Trizol reagent (Invitrogen, USA). Reverse transcription of RNA into cDNA was performed using a cDNA reverse transcription kit (11119ES60, Yeasen, China). qRT-PCR was performed on an ABI7500 PCR machine (ABI, USA) using the Hieff® Fast Cell Direct SYBR Green RT-qPCR Kit (11172ES40, Yeasen, China), with GAPDH as the internal reference. All samples were tested 3 times and changes in expression of target genes relative to GAPDH were calculated according to threshold cycle (Ct) analysis. Table 1 shows qRT-PCR primer sequences for HOXB9, CTHRC1, and GAPDH.
Table 1 The qRT-PCR primers sequences
| Primer | Forward | Reverse |
|---|---|---|
| CTHRC1 | 5'-ATGGGTACTAAACTGACTCAAC-3' | 5'-TCATTTTGGAAGCTCTTCAAT-3' |
| HOXB9 | 5'-CCATTTCTGGGACGCTTAGCA-3' | 5'-TGTAAGGGTGGTAGACGGACG-3' |
| GAPDH | 5'-GGACCTGACCTGCCGTCTAG-3' | 5'-GTAGCCCAGGATGCCCTTGA-3' |
qRT-PCR: quantitative real time-polymerase chain reaction.
Cell viability assay
Cells were collected 36-48 h after transfection and seeded into 96-well plates at the density of 1 × 103 cells/well. 10 μL of CCK-8 reagent (Beyotime, China) was supplemented for viability evaluation. Optical density at 450 nm was measured with a microplate reader after 2 h of culture. Incubation times were 0, 24, 48, and 72 h with three replicates per group.
Western blot
Western blot was performed in line with the protocols mentioned by Zhao et al.25. Extracted protein was submitted to BCA kit (P0010S, Beyotime, China) to measure protein concentration. SDS-PAGE electrophoresis was then performed. Proteins were transferred onto PVDF membranes (FFP26, Beyotime, China) and then blocked with 5% skimmed milk for 2 h, followed by overnight incubation with the primary antibodies at 4°C. After TBST (ST673, Beyotime, China) washes, proteins were incubated with the secondary antibody for 4 h at room temperature. Proteins were visualized with a high-sensitivity enhanced chemiluminescence (ECL) kit (P0018M, Beyotime, China) after washing with TBST. Finally, images were obtained through the chemiluminescence imaging system. The primary antibodies included rabbit anti-human FASN (ab128870), ACC1 (ab269273), acyl-CoA oxidase 1 (ACOX1) (ab184032), and GAPDH (ab181602). The secondary antibody was goat anti-rabbit IgG (ab205718). Primary and secondary antibodies were purchased from Abcam (UK).
Dual-luciferase reporter assay
Mutant and wild-type CTHRC1 promoters were cloned into the pGL3-Basic luciferase reporter vector (GenePharma, China). After that, pGL3-Basic-CTHRC1-promoter-WT and pGL3-Basic-CTHRC1-promoter-MUT plasmids were cotransfected into 293T cells with sh-HOXB9/sh-NC, respectively. 36-48 h after transfection, cells were collected and incubated with reporter lysis buffer (Promega, USA). Luciferase activity was measured by the Dual-Luciferase Assay System (Promega, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed in accordance with the protocols described by Tang et al.26. Antibodies to precipitate chromatin DNA were anti-HOXB9 (SCBT, USA) and antibodies used for immunoprecipitation controls were IgG (ab205718, Abcam, UK). qRT-PCR was performed to detect enrichment of CTHRC1. ChIP primers were as follows: forward primer 5'-GTTGTTATGGGTGTGTGGCAAGA-3', reverse primer 5'-TCCTGGGCTACTCTCATAAACG-3'.
Angiogenesis assay
50 μL of Matrigel (BD Biosciences, USA) was supplemented to each well of a 96-well culture plate and polymerized for 30 min at 37°C. Human umbilical vein ECs (HUVECs) seeded into Matrigel at a density of 2 × 104 cells/well were co-cultured with LUAD cell culture medium from different treatment groups. After incubation for 6 h at 37°C with 5% CO2, IX71 inverted microscope was used for observation27. The experimental results were counted according to the number of branches.
Quantitative analysis of metabolites
The lipophilic fluorescent dye BODIPY 493/503 (Invitrogen, USA) was utilized to detect neutral lipid accumulation in H1975 cells27.
Free FA and glycerol levels in cells were measured to assess lipolytic activity using EnzyChrom Free Fatty Acid Assay Kit (BioAssay Systems, USA) and Glycerol Cell Assay Kit (Cayman Chemical, MI), respectively28.
Analysis of statistics
All data were presented as mean ± standard deviation (SD) and all experiments were replicated three times. Statistical analysis was performed using SPSS statistical software 20.0 (IBM, USA). Normally distributed statistics were performed first, followed by t-test to analyze comparison between two groups, and one-way analysis of variance to analyze comparison among multiple groups. Correlations were analyzed by Pearson analysis. p < 0.05 suggested statistically significant.
RESULTS
CTHRC1 is highly expressed in LUAD
As bioinformatics analysis suggested that CTHRC1 was upregulated in LUAD tissues (Figure 1A, p < 0.001), its expression was assessed in a normal human lung epithelial cell line (BEAS-2B) and three different LUAD cell lines (A549, NCI-H1975, and PC-9). According to the results, higher expression of CTHRC1 was found in A549, NCI-H1975 and PC-9 (Figure 1B, p < 0.05). The above observations confirmed that CTHRC1 was substantially upregulated in LUAD. We concluded that CTHRC1 was highly expressed in LUAD cells.
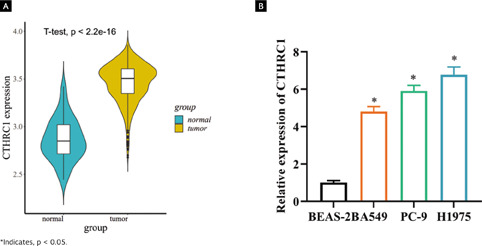
Figura 1. CTHRC1 is highly expressed in LUAD tissues. (A) Differentially expressed CTHRC1 in normal and tumor tissues was predicted by TCGA database. Blue violin indicates normal samples and yellow violin indicates tumor samples; (B) Expression of CTHRC1 mRNA in LUAD cell lines and normal human lung epithelial cell lines was detected by qRT-PCR.
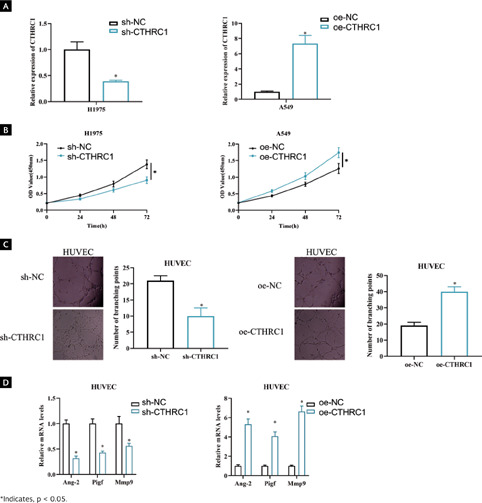
CTHRC1 promotes angiogenesis in LUAD cells
It has been documented that high CTHRC1 level may be strongly associated with tumor angiogenesis in LUAD29. To investigate the biological role of CTHRC1 in LUAD, we performed knockdown experiments on a cell line with high CTHRC1 expression (H1975) and overexpression experiments on a cell line with lower CTHRC1 expression (A549). qRT-PCR RIC version V2 revised results showed that CTHRC1 expression was significantly decreased on H1975 transfected with sh-CTHRC1 (p < 0.05), whereas a substantially increased CTHRC1 expression was observed on A549 transfected with oe-CTHRC1 when compared to the control (Figure 2A, p < 0.05). CCK-8 assay displayed a significant decrease in viability in CTHRC1 knockdown H1975 cells compared to control, and A549 cell viability was significantly enhanced upon overexpression of CTHRC1 (Figure 2B, p < 0.05). To investigate the influence of CTHRC1 on angiogenesis, we cocultured sh-CTHRC1/sh-NC and oe-CTHRC1/oe-NC treated cells with HUVECs cells, respectively, to detect angiogenic ability. The results of angiogenesis assay showed that knockdown of CTHRC1 expression suppressed the number of HUVECs branches and overexpression of CTHRC1 promoted the number of HUVECs branches compared to the control (Figure 2C, p < 0.05). Expression of genes involved in angiogenesis was analyzed by qRT-PCR. It was showed that knockdown of CTHRC1 suppressed mRNA levels of angiogenesis-related genes Ang-2 (angiopoietin), Pigf (placental growth factor), and Mmp9 (matrix metalloproteinase-9), while overexpression of CTHRC1 caused the opposite results (Figure 2D, p < 0.05). In summary, CTHRC1 played a role in promoting angiogenesis in LUAD.
CTHRC1 promotes Angiogenesis in LUAD cells by regulating FA metabolism
To explore the signaling pathway related to CTHRC1, we used GSEA database for prediction and found that CTHRC1 was enriched on different signaling pathways. Since we were interested in FA metabolic signaling pathways, we chose it as research object (Figure 3A). FAs are important cellular energy providers, important sources of triglycerides, and are closely associated with energy metabolism27. To test the hypothesis that CTHRC1 may be involved in FA metabolic reprogramming in LUAD, we generated cell groupings: oe-NC + DMSO, oe-CTHRC1 + DMSO, oe-RIC version V2 revised CTHRC1 + Orlistat (fatty acid inhibitor). Because CTHRC1 expression was relatively low in A549 cells, we transfected the above vectors into A549 for validation. First, we performed cell viability assays and the data showed that overexpression of CTHRC1 enhanced cell viability and Orlistat attenuated the effect of overexpression of CTHRC1 on cell viability (Figure 3B, p < 0.05). The effect of CTHRC1 on lipid content in LUAD cell lines was next examined. Cell staining was performed using the lipophilic fluorescent dye BODIPY 493/503 to detect levels of intracellular neutral lipids. The data showed that overexpression of CTHRC1 increased neutral lipids levels in A549 cells, whereas Orlistat reversed the effect of overexpression of CTHRC1 on neutral lipid levels (Figure 3C, p < 0.05). In addition, we examined intracellular levels of FAs and glycerol to assess lipolytic activity. The results indicated that intracellular FA and glycerol levels were notably increased in A549 cells after overexpression of CTHRC1, whereas Orlistat reversed the effect of overexpression of CTHRC1 on FAs and glycerol levels (Figure 3D, p < 0.05). To demonstrate the role of CTHRC1 in regulating FAs metabolism in LUAD, we examined the effect of CTHRC1 on several other key FAs metabolizing enzymes in LUAD cells, including acetyl-CoA carboxylase α (ACC1), fatty acid synthase (FASN), and ACOX1. The results showed that FASN and ACC1 protein levels were significantly up-regulated and ACOX1 protein levels were down-regulated in A549 cells after overexpression of CTHRC1 compared with controls; while Orlistat reversed the effect of overexpression of CTHRC1 on key enzymes of FA metabolism (Figure 3E, Supplementary Figure 1A, p < 0.05). The above results indicated that CTHRC1 promoted the reprogramming of FA metabolism in LUAD.
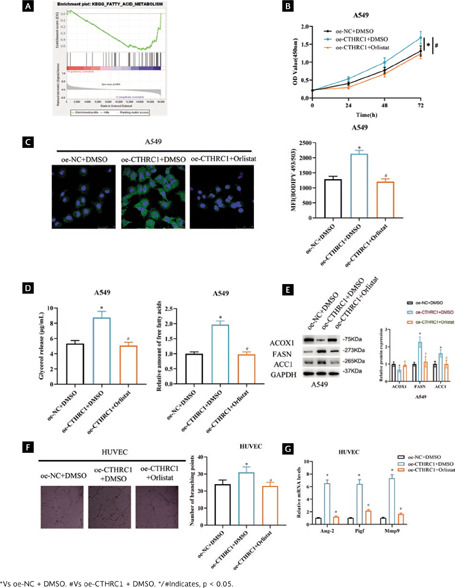
Figura 3. CTHRC1 promotes angiogenesis in LUAD cells by regulating FA metabolism. (A) GSEA enrichment pathway map of CTHRC1; (B) LUAD cell viability after transfection by CCK-8; (C) Lipid accumulation detected by metabolite quantitative analysis assay after transfection, MFI: mean fluorescence intensity; (D) FAs and glycerol levels in LUAD cells after transfection treatment were detected; (E) Expression of FA key metabolic enzymes after transfection; (F) Angiogenic ability after transfection; (G) Expression of angiogenesis-related genes after transfection.
FA composition has been shown to modulate the angiogenic response30,31. To verify whether CTHRC1 promotes angiogenesis in LUAD cells by regulating the FAs metabolic pathway. We performed angiogenesis assays and showed that overexpression of CTHRC1 promoted branch formation in RIC version V2 revised HUVECs compared to control, whereas Orlistat reversed the effect of overexpression of CTHRC1 on branch formation in HUVECs (Figure 3F, p < 0.05). Subsequently, qRT-PCR analysis showed that mRNA levels of Ang-2, Pigf, and Mmp9 were significantly increased upon overexpression of CTHRC1, and Orlistat reversed the effect of overexpression of CTHRC1 on angiogenesis-related gene expression (Figure 3G, p < 0.05). The above findings suggested that CTHRC1 promoted angiogenesis in LUAD cells by regulating FA metabolism.
HOXB9 is a transcription factor upstream of CTHRC1
To find out more about the gene affecting CTHRC1 in LUAD, 90 potential CTHRC1 transcription factors returned from the KnockTF database were intersected with 90 transcription factors and 2-fold up-regulated differential genes, which yielded four candidate genes (TWIST1, FOXM1, HMGA1 and HOXB9). TWIST1, FOXM1, and HMGA1 have been well studied in lung cancer, but HOXB9 has not been studied in depth, so we chose HOXB9 as the target transcription factor of CTHRC1 (Figure 4A). Among them, the correlation between HOXB9 and CTHRC1 was approximately 0.2, which exhibited a significant positive correlation (p < 0.05) (Figure 4B, Supplementary Table 1). Bioinformatics analysis uncovered that HOXB9 expression was upregulated in LUAD tissues (Figure 4C, p < 0.001). qRT-PCR results showed that HOXB9 expression was significantly upregulated in different LUAD cell lines (Figure 4D, p < 0.05). We predicted the motif of HOXB9 by the Jaspar database to score up to 8.2 points for affinity at 2000 bp upstream of CTHRC1, with possible binding sites (Figure 4E). Dual luciferase reporter assay and ChIP assay were used to verify the binding relationship between HOXB9 and CTHRC1. The results of dual luciferase assay showed that sh-HOXB9 markedly inhibited the luciferase activity of CTHRC1-WT (Figure 4F, p < 0.05). ChIP assay confirmed that anti-HOXB9 significantly promoted enrichment of CTHRC1 (Figure 4G, p < 0.05). Since the above results showed that HOXB9 could bind the promoter region of CTHRC1, HOXB9 was selected as a candidate transcription factor for subsequent experiments.
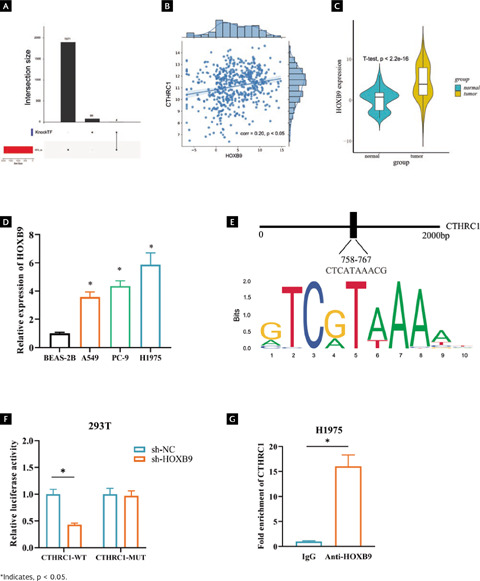
Figura 4. HOXB9 is a transcription factor upstream of CTHRC1. (A) Upsetplot plots of CTHRC1 upstream transcription factors from KnockTF database; (B) A significant positive correlation between CTHRC1 and HOXB9 by Pearson analysis; (C) Predicted differential expression of HOXB9 in normal and LUAD tissues was predicted by TCGA database; (D) Differential expression of HOXB9 in normal cells BSEA-2B and LUAD cells A549, PC-9 and H1975; (E) Binding of CTHRC1 and its upstream transcription factor HOXB9; (F) and (G) Binding relationship between HOXB9 and CTHRC1 validated by dual luciferase assay and ChIP assay.
Activation of CTHRC1 by HOXB9 promotes angiogenesis in LUAD cells through FA metabolism
To further study the function and role of HOXB9 and CTHRC1 in LUAD. We constructed sh-NC+oe-NC, sh-HOXB9+oe-NC, and sh-HOXB9+oe-CTHRC1 vectors to transfect H1975 cells, respectively. When cell transfection efficiency was measured by qRT-PCR, knockdown of HOXB9 dramatically decreased CTHRC1 expression, whereas overexpression of CTHRC1 reversed the effect of knockdown of HOXB9 (Figure 5A, p < 0.05). CCK-8 results showed that cell viability of H1975 was significantly reduced after knockdown of HOXB9, whereas overexpression of CTHRC1 offset the effect of knockdown of HOXB9 expression on cell viability (Figure 5B, p < 0.05). To investigate how HOXB9 affects FA metabolism pathways, first we examined the effect of each treatment group on the lipid content of the H1975 cell line by staining cells with the lipophilic fluorescent dye BODIPY 493/503. The results showed that HOXB9 knockdown decreased the level of neutral lipids in H1975 cells, whereas overexpression of CTHRC1 reversed the effect of HOXB9 knockdown on neutral lipid levels (Figure 5C, p < 0.05). Subsequently, to assess lipolytic activity, we examined intracellular levels of FAs and glycerol. The results showed that knockdown of HOXB9 decreased the levels of FAs and glycerol in H1975 cells, whereas overexpression of CTHRC1 reversed the effect of knockdown of HOXB9 on FAs and glycerol levels (Figure 5D, p < 0.05). In addition, we examined the effect of HOXB9 on key FA metabolizing enzymes (FASN, ACOX1, and ACC1) in LUAD cells. The results showed that knockdown of HOXB9 decreased protein levels of FASN and ACC1 and increased ACOX1 protein level compared to controls. Whereas overexpression of CTHRC1 reversed the effect of knockdown of HOXB9 on key enzymes of FA (Figure 5E and Supplementary Figure 1B, p < 0.05). The above results indicate that HOXB9 promotes FA metabolic processes in LUAD by activating CTHRC1. In the following part, we explored the effect of HOXB9 on angiogenesis. The results of the angiogenesis assay showed that knockdown of HOXB9 inhibited branching of HUVECs compared to control, whereas overexpression of CTHRC1 reversed the influence of knockdown of HOXB9 on branching of HUVECs (Figure 5F, p < 0.05). Finally, qRT-PCR was used to detect expression of angiogenesis-related genes. It was found that knockdown of HOXB9 significantly inhibited the mRNA levels of Ang-2, Pigf, and Mmp9, and overexpression of CTHRC1 reversed the effect of HOXB9 knockdown on angiogenesis-related gene expression (Figure 5G, p < 0.05). The above results indicate that activation of CTHRC1 by HOXB9 promotes angiogenesis in LUAD cells through FA metabolism pathway.
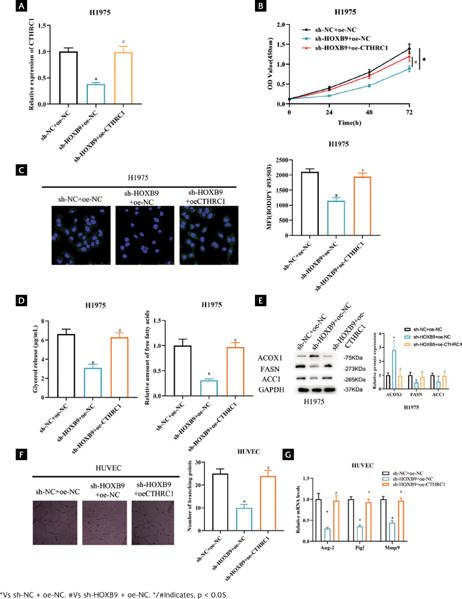
Figura 5. Activation of CTHRC1 by HOXB9 promotes angiogenesis in LUAD cells through FA metabolism. (A) Transfection efficiency; (B) LUAD cell viability after transfection; (C) Lipid accumulation after transfection; MFI: mean fluorescence intensity; (D) FAs and glycerol levels in LUAD cells after transfection; (E) Expression of key FA metabolizing enzymes after transfection; (F) Angiogenesis ability after transfection; (G) Expression of angiogenesis-related genes after transfection.
DISCUSSION
By far, LUAD remains the most invasive and fatal cancer. Although numerous therapies are in place to treat LUAD, its high heterogeneity and complex molecular mechanisms32 often bring about unsatisfactory outcomes in clinics. Therefore, the urgent task is to unveil the molecular pathology of LUAD and identify relevant therapeutic targets, hopefully resulting in tangible benefits in LUAD treatment. CTHRC1 participates in physiological processes like angiogenesis and bone formation10,33. For example, Fu et al.34 confirmed that CTHRC1 induced migration and angiogenesis of HUVECs in GIST by elevating phosphorylation of extracellular signal-regulated protein kinase (ERK) and c-Jun N-terminal kinase (JNK) signaling pathways. CTHRC1 has been reported to be highly expressed in multiple tumors with a cancer-promoting role35-37. As demonstrated by Ni et al.36, CTHRC1 facilitates colorectal cancer cell viability and EMT through activation of the transforming growth factor-beta (TGF-β) signaling pathway. This study confirmed that overexpression of CTHRC1 could promote tumor angiogenesis and cell viability thereby promoting LUAD progression. However, knockdown of CTHRC1 could repress tumor angiogenesis and cell viability. Our study provides a promising therapeutic target for anti-angiogenic therapy.
Herein, we found the upstream transcription factor HOXB9 in CTHRC1 by bioinformatics analysis, and confirmed that HOXB9 could activate the transcription of CHRC1 by molecular experiments. The HOX gene was originally described as a developmental gene, which encodes key transcription factors that exert vital functions in embryogenesis19. Expression of the HOX gene complex directly contributes to tumor progression by repressing apoptosis, altering receptor signaling, and facilitating EMT and invasion38. As the ninth analogous locus in the HOX-B cluster, HOXB9 is involved in the growth of a variety of cancers. However, due to tumor heterogeneity, HOXB9 may be an oncogene or tumor suppressor in different cancers. For instance, HOXB9 is a gastric cancer suppressor, whose overexpression increases gastric cancer cell apoptosis and inhibits proliferation and metastasis39. In colon and lung cancer, HOXB9 plays a cancer-promoting role. Hoshino et al.40 showed that HOXB9 could induce the secretion of angiogenic factors by IL6 signaling activation, promoting proliferation and colorectal cancer progression. Zheng et al.41 showed that in LUAD, HOXB9 detaches NSCLC cells from the primary tumor by inducing EMT and accelerates brain metastasis by driving MMP9 production and degradation of intercellular adhesion proteins in ECs of the blood-brain barrier. In this study, HOXB9 was proved to be oncogenic in LUAD as it promotes LUAD cell angiogenesis by activating CTHRC1. These observations add up to suggest that HOXB and CTHRC1 may be potential therapeutic targets to inhibit the progression of LUAD.
Reprogramming of cellular energy metabolism is widely recognized as a novel hallmark of cancer28. FA metabolism is a novel feature of malignancy that consists of anabolism and catabolism that maintain energy homeostasis. A loss of balance between FA synthesis and breakdown may lead to inadequate FA levels or lipid accumulation42. At present, the role of FA metabolism reprogramming in the progression of LUAD has received tons of attention. For example, Garcia et al.43 found that FA binding protein 5 (FABP5) is involved in FA synthesis. FABP5 expression not only promotes the expression of FA synthesis-related enzymes such as FASN and SCD1 but also is necessary for migration, cell cycle progression and tumor growth in vivo in LUAD. As a rate-limiting enzyme of unsaturated FA synthase, SCD1, which is highly expressed in LUAD, is necessary for cell proliferation, migration, and invasion. Patients with advanced LUAD tend to have increased SCD1 expression and a poor prognosis44. We demonstrated that CTHRC1 could promote LUAD cell activity through FA metabolism. FA metabolism also plays an imperative regulatory role in tumor angiogenesis, as Chen et al.27 found that CCAT1 promotes cell angiogenesis and proliferation by modulating FA metabolism in LUAD. Sun et al.45 showed that PKM2 promotes LUAD cell invasion and proliferation as well as angiogenesis by activating glycolytic and lipid synthesis pathways. In this study, HOXB9 was discovered to promote angiogenesis in LUAD cells by activating CTHRC1 to regulate the fatty acid metabolic pathway. Our results suggested that CTHRC1 and HOXB9 were important regulatory molecules in LUAD cell metabolism, suggesting that the CTHRC1/HOXB9 axis may be an important target for inhibiting LUAD lipid metabolism pathways and also offer a new strategy for LUAD treatment based on metabolic reprogramming pathways.
In summary, this study confirmed the molecular mechanism that HOXB9 targeted CTHRC1 to regulate the FA metabolic pathway to promote angiogenesis in LUAD cells. These findings provide new perspectives for the investigation of LUAD mechanisms. However, one limitation of this study is that the finding has not been validated in vivo in the animal experiment. Our team will continue to explore the pro-angiogenic effects of the HOXB9/CTHRC1 axis in the future experiments in mice. In conclusion, our study was a first approach that suggest that CTHRC1 could be a target molecule for the LUAD treatment.
SUPPLEMENTARY MATERIAL
Supplementary data are available at Revista de Investigación Clínica online (10.24875/RIC.23000023). These data are provided by the corresponding author and published online for the benefit of the reader. The contents of supplementary data are the sole responsibility of the authors.











 text new page (beta)
text new page (beta)