Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ingeniería química
versão impressa ISSN 1665-2738
Rev. Mex. Ing. Quím vol.10 no.1 Ciudad de México Abr. 2011
Biotecnología
Characterization of an aspartic protease produced by Amylomyces rouxii
Caracterización de una aspartato proteasa producida por Amylomyces rouxii
J. Marcial1, A.I. Pérez de los Santos1, F.J. Fernández1, G. Díaz–Godínez2, A.M. Montiel–González2 and A. Tomasini1*
1 Depto. de Biotecnología, Univ. Autónoma Metropolitana–Iztapalapa. Apdo. Postal 55–535, C.P. 09340, México, DF, México. *Corresponding author. E–mail: atc@xanum.uam.mx Tel. (+52–55) 58046453.
2 Universidad Autónoma de Tlaxcala. Centro de Investigación en Ciencias Biológicas. Tlaxcala, Tlaxcala, C.P. 90070, México.
Received 11 of October 2010.
Accepted 20 of January 2011.
Abstract
The zigomycete Amylomyces rouxii was isolated, in our laboratory, from effluents of paper industries, it was showed that this fungus produces extracellular tyrosinase. In this work, we show that this fungus produces also an extracellular protease. The extracellular protease was partially purified using two–step purification, by (NH4)2SO4 fractionation and DEAE–sepharose anion exchange chromatography with 4.25–fold factor purification. The optimal pH for protease activity, using hemoglobin as substrate, was 3.5. Results from protein purification revealed that this protease is an aspartic protease. Protease activity was inhibited 73% by pepstatin A, a specific inhibitor of acid proteases. Results allow conclude that A. rouxii produce an extracellular aspartic protease that has similar characteristics to aspartic proteases produced by fungi as Mucor and Rhizopus.
Keywords: Amylomyces rouxii, aspartic protease, inhibitor protease.
Resumen
Amylomyces rouxii es un zigomiceto que fue aislado, en nuestro laboratorio, a partir de un efluente de la industria del papel, este hongo produce tirosinasa extracelular. En este trabajo demostramos que además de la tirosinasa A. rouxii produce una proteasa extracelular. Esta proteasa se purificó parcialmente en dos pasos, por precipitación con (NH4)2SO4 y después se pasó a través de una columna de intercambio aniónico, el factor de purificación obtenido fue de 4.25. Su pH óptimo usando hemoglobina como sustrato fue de 3.5. Los resultados de la purificación de la proteína demuestran que se trata de una aspartato proteasa, la actividad proteasa fue inhibida en 73% por pepstatin A, un inhibidor específico para proteasas tipo ácido. Los resultados permiten concluir que A. rouxii produce una aspartato proteasa extracelular con características similares a las aspartato proteasas producidas por los hongos del género Mucor y Rhizopus.
Palabras clave: Amylomyces rouxii, aspartato proteasa, inhibidor de proteasa.
1 Introduction
Amylomyces rouxii is a zigomycete isolated from effluents of paper industries (Tomasini et al., 1996). A. rouxii has a high similarity with Rhizopus oryzae (Montiel et al., 2004; Kito et al., 2009). This fungus produces tyrosinase (Montiel et al., 2004). Tyrosinase is used in food, cosmetic and pharmaceutical industries (Seo et al., 2003; Halaouli et al., 2006; Selinheimo et al., 2006) and also has been used in bioremediation process of waste and soil containing phenolic compounds (Montiel et al., 2004; Girelli et al., 2006; Kim et al., 2008).
On the other hand, many authors have reported fungal proteases produced by Mucor and Rhizopus, as it is known proteases are one of groups of enzyme most important in industrial processes. The fungal proteases are mainly proteases type acid and can substitute the role of three important protease involved in food processing, i.e. pepsin, rennin and papain. Acid proteases are the main type of protease secreted by Mucor and Rhizopus spp. Aspartic protease from Mucor miehei, M. rennin and M. pusillus have been studied (Gray et al., 1986; Etoh et al., 1982; Tonouchi et al., 1986). Also some authors have reported the purification and characterization of aspartic proteases from Rhizopus microsporus var rhizopodiformis, R. chinesis, R. hangchow, R. oryzae, Mucor sp, and Rhizomucor miehei (Preetha and Boopathy, 1997; Fernández–Lahore et al., 1999; Kumar et al., 2005; Chen et al., 2009). Aspartic proteases have been used as calf rennet substitutes in cheese production. This is a chief enzyme employed in cheese production, aspartic protease not only clots the milk but also play an important role during cheese maturation, which is a vital and complex process for the balanced development of flavor and texture (Fox et al., 1993; Vioque et al., 2000; Fernández–Lahore et al., 1999; Kumar et al., 2005). An aspartic protease from Trichoderma harzianum has been identified as a cell wall–degrading enzyme involved in biocontrol activities (Suárez et al., 2005; Liu and Yang, 2007).
The aim of this work was to know if A. rouxii also produces some proteases, as well as to partially purify and characterize one of these proteases produced by A. rouxii.
2 Methodology
2.1 Strain, media, and growth conditions
A. rouxii, isolated from effluents of paper industries (Tomasini et al, 1996), was used in this study. The strain was maintained by freezing spores suspension at 40% of glycerol at — 20° C.
Erlenmeyer flasks containing (potato agar dextrose) PDA were inoculated and incubated at 30° C for 4 days to obtain spores. Spore suspension was obtained in 20 ml of sterile water containing 0.1% Tween–80 and it was used to inoculate the submerged culture for enzymes production. Cultures were performed in flasks containing 50 mL of modified Melin–Norkrans medium containing (g L–1): glucose 5, malt extract 2, yeast extract 1, KH2PO4 0.5, MgSO4·7H2O 0.15, (NH 4)2HPO 4 0.5, pH 5.6. In this study, 0.1 g tyrosine L–1 and 0.0125 g pentachlorophenol L–1 were added to the medium, as reported by Montiel et al, (2004) who showed that these compounds increased tyrosinase activity. Medium was inoculated with 1 x 106 spores mL–1, and incubated at 30°C in a rotary shaker at 150 rpm (Tomasini et al, 2001). The enzymes were harvested after 48 h of incubation.
2.2 Preparation of extracellular extract
Crude enzyme extract was obtained from 500 mL of broth culture from A. rouxii at 48 h. The culture medium was separated from the mycelium by filtration through Whatman filter paper 41; an aliquot of this crude extract was used to determine protease activity. The rest of the extract was frozen at —80°C and lyophilized. The lyophilized sample from broth cultures was suspended in 30 mL of 10 mM phosphate buffer pH 6.8. This extract was used for enzymatic activity determination, protein quantification, as well as for the partial purification and characterization of the extracellular protease. To conduct the inhibition studies the following substances were used: 1 mM PMSF, 1 mM EDTA, 3 mM pepstatin A and a commercial cocktail (200 pL concentration, according to sigma) that is a mixture of protease inhibitors, containing 4–2–aminoethyl–benzenesulfonyl fluoride, pepstatin A, E–64, bestatin, leupeptin and aprotinin (sigma, P8340).
2.3 Partial purification of extracellular p rotease
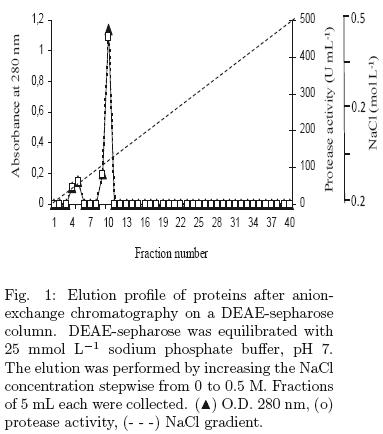
Proteins from the crude extract were precipitated with ammonium sulphate to 80% w/v saturation. The crude extract was then centrifuged at 20,000 g, for 30 min, at 4°C. The proteins were dialyzed against milli–Q water for 24 h using a 10–kDa membrane cut–off, with three repeated changes of water. Samples containing proteins were poured into an anion–exchange chromatographic column (DEAE–Sepharose, Amersham) equilibrated beforehand with 25 mmol L–1 sodium phosphate buffer, pH 7. Proteins were eluted with a linear gradient of 0–0.5 M NaCl in 25 mmol L–1 sodium phosphate buffer, pH 7, at a flow rate of 1 mL min–1. Fractions of 5 mL were collected, and the protein concentration and protease activity was determined in each fraction. Fractions with enzymatic activities were concentrated using Microcon tubes YM–10 (0.5 mL Millipore).
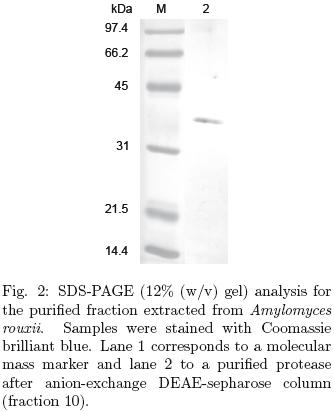
The purity of proteins from fractions showing protease activity was analyzed in denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), performed with 10% acrylamide/gels according to Laemmli (1970). The gels were stained using Coomasie brilliant blue R–205 (Sigma) for 30 min followed by incubation over–night with destaining solution.
The protein of each fraction was monitored by measuring the absorbance at 280 nm. Protein concentrations were determined by the Bradford method (Bradford, 1976) using bovine serum albumin as standard.
2.4 Protease activity assay
The protease activity was determined according to the methods described by Anson (1938) and Asakura et al. (1997). Hemoglobin (2% w/v; Sigma, USA) dissolved in universal buffer, pH 3.5, and azocasein (1% w/v; Sigma, USA) dissolved in 0.2 M citrate buffer, pH 7.5, were used as substrates. Enzyme extract (0.25 mL) was added to 1 mL of substrate and incubated at 37°C in a water bath for 20 min. The reaction was stopped by adding 0.25 mL trichloroacetic acid (10% w/v) and centrifuged at 20,000 g for 15 min to remove the precipitate. When azocasein was used, one unit of protease activity was defined as the amount of enzyme required to liberate 1.0 μmol of tyrosine per minute at 37°C. For hemoglobin as substrate, one unit of protease activity was the amount necessary to produce an increase in absorbance of 0.01 at 280 nm after 20 min.
3 Results and discussion
3.1 Partial purification of the protease
Protease activity from extract crude, using azocaseine as substrate, was 0.706 ± 0.77 U mL–1 and, with hemoglobin, it was 273.86 ± 7.16 U mL–1. Protease activity was 387–times higher when hemoglobin was used as substrate at pH 3.5 than when using azocasein as substrate at pH 7.0. Then, protease activities reported in the following results are determined only using hemoglobin as substrate.
The crude enzyme extract from the lyophilized sample was precipitated using 80% (NH4)2SO4 and the enzyme was further partially purified by anion–exchange chromatography, DEAE–Sepharose, the procedure is summarized in Table 1. A single peak of protease activity was eluted in fraction 10, (Table 1, Fig. 1) with a 4.25–fold purification. This value of purification factor is similar to the value reported by an aspartic protease from Ce ntaurea calci trapa, obtained in the same purification step (Raposo and Domingos 2008). The molecular mass of the purified enzyme was approximately 40 kDa by SDS–PAGE (Fig. 2). These results strongly suggest that the protease produced by A. rouxii is an acid protease.
3.2 Identification of protein and amino acid sequence
Peptides sequence was attained by electrospray LC–MS/MS (IBT, UNAM). The N–terminal amino acid sequence of the purified protease showed 99% similarity to rhizopuspepsin–2 precursor (aspartate protease) of Rhizopus oryzae. These results confirm that the extracellular protease produced by A. rouxii is an acid protease, specifically an aspartic protease.
Based on its similitude with other fungal aspartic protease, the protease from A. rouxii was expected to contain 391 residues, with a molecular weight of 40 kDa.
Analysis of the amino acid sequence by SignalP V3.0 identified a cleavage signal–sequence site between positions TEA21 and A22. The putative signal peptide corresponding to the first 21 amino acid shows typical features of signal peptides, such as a highly hydrophobic region and alanine residues at –3 and –1 position relative to the cleavage site (Nielsen et al., 1997).
The amino acid sequence found in A. rouxii has a consistent degree of similarity with aspartate proteases, according to BlastP analysis (GenBank non–redundant protein sequences). The highest sequence identity was with the aspartic protease from Rhizopus oryzae (99%). Lower identity percentages were found with the aspartic protease from Aspergillus oryzae (40%), Fusarium venenatum (39%), Hypocrea jecorina (37%), and Lentinula edodes (32%), data shown in Fig. 3. In fungal aspartic proteases, the catalytic residue is contained within the conserved motif DTGS and DTGT as was found in the protease sequence from A. rouxii (Fig. 3). The residues involved in substrate specificity were present at positions Y144, G145, and D146, which is conserved in fungal aspartic protease (Fig. 3); hydrophobic motifs responsible for substrate specificity were found also at positions LLD191 and IFD367. Another important feature is related to disulfide bonds, the aspartic protease of mammalian origin contains three characteristic loops, the first loop is present in several fungal enzymes, such as Trichoderma harzianum (cystein residues C99 and C105) (Suárez et al., 2005) and in R. oryzae and A. rouxii at C115 and C118 positions. A second loop at C319 and C352 is also conserved in A. rouxii and R. oryzae.
3.3 Characterization of the aspartic protease from A. rouxii
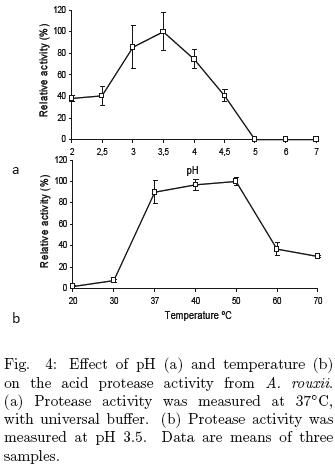
The effect of pH on protease activity was studied using hemoglobin as substrate. The aspartic protease from A. rouxii was active in the pH range 2–4.5, with an optimal pH of 3.5 (Fig. 4a), which is similar to the acid protease from Aspergillus oryzae MTCC 5341 (Vishwanatha et al., 2009) and to the rhizopuspepsin from Rhizopus oryzae NBRC 4749 (Chen et al., 2009). Maximal protease activity from A. rouxii was found in the temperature range from 37 to 50°C (Fig. 4b). The optimal temperature reported for the acid protease from Aspergillus oryzae MTCC 5341 is 55°C (Vishwanatha et al., 2009) and for the rhizopuspepsin from Rhizopus oryzae NBRC 4749 is 50°C (Chen et al., 2009). The activation energy of aspartic protease was 55.82 kcal mol–1; this value was calculated from the data obtained of Fig. 4b using Arrhenius equation.
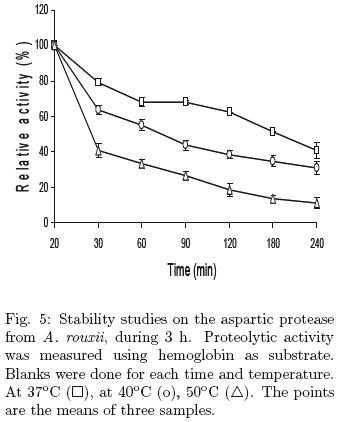
The thermal stability assays showed that the aspartic protease at 37°C retained 70% of the initial activity after 1 h incubation, and this activity was similar after 1 h, while 60 and 30% of the initial activity remained within 1 h of incubation at 40 and 50°C (Fig. 5). Similar thermal stability has been reported for Rhizopus oryzae and Aspergillus oryzae (Chen et al., 2009, Vishwanatha et al., 2009).
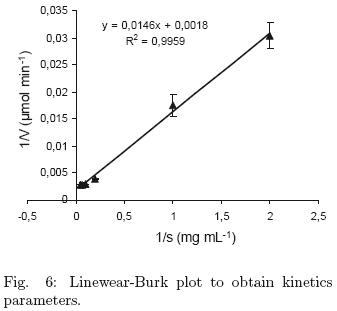
The Km and Vmax values for the partial purified protease were determined using 0 – 20 mg mL–1 hemoglobin as substrate. Km value was 8.03 mg mL–1 and Vmax was 551.7 μmol min–1, calculated from Linewear–Burk plot (Fig. 6).
3.4 Effect of inhibitors
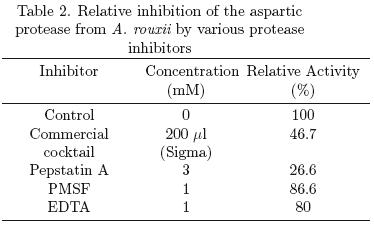
Various protease inhibitors were assayed and results showed that pepstatin A (3 mM) inhibited 73% the aspartic protease activity. It has been reported that pepstatin A is specific inhibitor for acid proteases, binding to the active aspartate site (Palmieri et al., 2001; Kudryavtseva et al., 2008). Kumar et al., (2005) reported an acid protease from R. oryzae and the inhibition of its activity was of 73 and 93% using 0.01 and 0.02 mM of pepstatin A, respectively. Vishwasnatha et al. (2009) observed 100% inhibition of acid protease from A. oryzae using 0.018 mM of pepstatin A. Both authors reported that PMSF did not inhibit acid protease activity, similar results were obtained in this work, PMSF inhibited only 13% of the aspartic protease activity (Table 2).
Conclusion
The results obtained indicate that A. rouxii besides produce tyrosinase, as has been reported, produce also an aspartic protease. This protease showed higher activity when hemoglobin as used as substrate that when azocasein was used. Protease produced by A. rouxii was identified as an aspartic protease. The protease from A. rouxii showed similar characteristics with other aspartic protease from Rhizopus. This study allows proposing A. rouxii as a fungus producer extracellular aspartic protease. The culture medium used was proposed to produce tyrosinase, it must be assayed other culture media in order to increase protease production. Also, it could be study the potential of this protease to be used in cheese production or whatever use.
Acknowledgements
J. Marcial and Pérez de los Santos were supported by fellowships, applications 172706 and 229059 respectively from CONACYT, México.
References
Anson, M.L. (1938). The estimation of pepsin, trypsin, papain and cathepsin with hemoglobin. Journal of General Physiology 22,79–89. [ Links ]
Asakura, T., Watanabe, H., Ab, K. and Arai, S. (1997). Oryzasin as an aspartic proteinase occurring in rice seeds: purification, characterization, and application to milk clotting. Journal of Agricultural and Food Chemistry 45, 1070–1075. [ Links ]
Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254. [ Links ]
Chen, C–C., Cho, Y.C., Lai, C–C. and Hsu, W–H. (2009). Purification and characterization of a new rhizopuspepsin from Rhizopus oryzae NBRC 4749. Journal of Agricultural and Food Chemistry 57, 6742–6747. [ Links ]
Etoh, Y., Shoun, H., Ogino, T., Fujiwara, S., Arima, K. and Beppu, T. (1982). Proton magnetic resonance spectroscopy of an essential histidyl residue in a milk–clotting acid protease, Mucor rennin. Journal of Biochemistry 91, 2039–2046. [ Links ]
Fernández–Lahore, H.M., Auday, R.M., Fraile, E.R., Biscoglio de Jiménez, B.M., Pripignani, L., Machalinski, C. and Cascone, O. (1999). Purification and characterization of an acid proteinase from mesophilic Mucor sp. solid–state cultures. Journal of Pepetide Research 53, 599–605. [ Links ]
Fox, P.F, Law, J, McSweeney P.L.H. and Wallace, J. (1993). Biochemistry of cheese ripening. In: Fox PF, editor. Cheese chemistry, Physics and Microbiology, vol. II. London: Chapman & Hall. P. 389–438. [ Links ]
Girelli, A.M., Mattei, E. and Messina, A. (2006). Phenols removal by immobilized tyrosinase reactor in on–line high performance liquid chromatography. Analytical Chemistry Acta 580, 271–277. [ Links ]
Gray, G.L., Hayenga, K., Cullen, D., Wilson, L.J. and Norton, S. (1986). Primary structure of Mucor miehei aspartic protease: evidence for a zymogen intermediate. Gene 48, 41–53. [ Links ]
Halaouli, S., Asther, M., Sigoillot, J.C., Hamdi, M. and Lomascolo, A. (2006). Fungal tyrosinases: new prospects in molecular characteristics, bioengineering and biotechnological applications. Journal of Applied Microbiology 100, 219–232. [ Links ]
Kim, G.Y., Shim, J., Kang, M.S. and Moon, S.H. (2008). Optimized coverage of gold nanoparticles at tyrosinase electrode for measurement of a pesticide in various water samples. Journal of Hazardous Material 165, 141–147. [ Links ]
Kito, H., Abe, A., Sujaya, I.N., Oda, Y., Asano, K. and Sone, T. (2009). Molecular characterization of the relationship among Amylomyces rouxii, Rhizopus oryzae, and Rhizopus delemar. Bioscience Biotechnology and Biochemistry 73, 861–864. [ Links ]
Kudryavtseva, O.A., Dunaevsky, Ya.E., Kamzolkina, O.V. and Belozersky, M.A. (2008). Fungal proteolytic enzymes: features of the extracellular proteases of xylotrophic basidiomycetes. Micobiology 77, 725–737. [ Links ]
Kumar, S., Sharma, N.S., Saharan, M.R. and Singh, R. (2005). Extracellular acid protease from Rhizopus oryzae: purification and characterization. Process Biochemistry 40, 701–1705. [ Links ]
Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [ Links ]
Liu, Y. and Yang, Q. (2007). Cloning and heterologous expression of aspartic protease SA76 related to biocontrol in Trichoderma harzianum. FEMS Microbiology Letters 277, 173–181. [ Links ]
Montiel, A.M., Fernández ,F.J., Marcial, J., Soriano, J., Barrios–González, J. and Tomasini, A. (2004). A fungal phenoloxidase (tyrosinase) involved in pentachlorophenol degradation. Biotechnology Letters 26, 1353–1357. [ Links ]
Nielsen, T.H., Deiting, U. and Sitt, M. (1997). A –amylase in potato is induced by storage at low temperature. Plant Physiology 113, 503–510 [ Links ]
Palmieri, G., Bianco, C., Cennamo, G., Giardina, P., Merino ,G., Monti, M. and Sannia, G. (2001). Purification, characterization, and functional role of a novel extracellular protease from Pleurotus ostreatus. Applied and Environmental Microbiology 67, 2754–2759. [ Links ]
Preetha, S. and Boopathy, R. (1997). Purification and characterization of a milk clotting protease from Rhizomucor miehei. World Journal of Microbiology and Biotechnology 13, 573–578. [ Links ]
Raposo, S. and Domingos, A. (2008). Purification and characterization milk–clotting aspartic proteinases from Centaurea cal citrapa cell suspension cultures. Process Biochemistry 43, 139–144. [ Links ]
Selinheimo, E., Saloheimo, M., Ahola, E., Westerholm–Parvinen, A., Kalkkinen, N., Buchert, J. and Cruz, K. (2006). Production and characterization of a secreted, C–terminally processed tyrosinase from the filamentous fungus Trichoderma reesei. The FEBS Journal 273, 4322–4325. [ Links ]
Seo, S.Y., Sharma, V.K. and Sharma, N. (2003). Mushroom tyrosinase: Recent prospect. Journal of Agricultural and Food Chemistry 51, 2837–2853. [ Links ]
Suárez, B., Sanz, L., Chamorro, I., Rey, M., González, F.J., Llobell, A. and Monte, E. (2005). Proteomic analysis of secreted proteins from Trichoderma harzianum identification of a fungal cell wall–induced aspartic protease. Fungal Genetics Biology 42, 924–934 [ Links ]
Tomasini, A., Villareal–Arellanos, H.R. and Barrios–González, J. (1996). Resistencia de una cepa de Rhizopus sp. al crecer en medios conteniendo pentaclorofenol. Avances en Ingeniería Química 6, 36–40. [ Links ]
Tomasini, A., Flores, V., Cortés, D. and Barrios–González, J. (2001). An isolate of Rhizopus nigricans capable of tolerating and removing pentachlorophenol. World Journal of Microbiology and Biotechnology 17, 201–205. [ Links ]
Tonouchi, N., Shoun, H., Uozumi, T. and Beppu, T. (1986). Cloning and sequencing of a gene for Mucor rennin, an aspartate protease from Mucor pusillus. Nucleic Acids Research 14, 7557–7569. [ Links ]
Vioque, M., Gomez, R., Sanchez, E., Mata, C., Tejeda, L., Fernandez–Salguero, J. (2000). Chemical and microbiological characteristics of ewe's milk cheese manufactured with extracts from flowers of Cynara cardunculus and Cynara humilis as coagulants. Journal of Agriculture and Food Chemistry 48, 451–456. [ Links ]
Vishwanatha, K.S., Appu–Rao, A.G. and Singh, S.A. (2009). Characterization of acid protease expressed from Aspergillus oryzae MTCC 5341. Food Chemistry 114, 402–407. [ Links ]














