Highlights
Fusarium wilt caused by Fusarium oxysporum f. sp. niveum (Fon) is a major fungal disease affecting watermelon crops worldwide.
This disease can result in losses of up to 100 % when cultivars not resistant to the fungus are used.
Three Fusarium isolates were obtained from symptomatic watermelon plants and they were identified as Fon race 1.
This is the first report where Fon race 1 is identified as the causal agent of watermelon wilt and vascular necrosis in Mexico.
Introduction
Mexico is one of the top 10 watermelon-producing countries in the world, with a production of 1,362,393 t and an area of 40,000 ha (Servicio de Información Agroalimentaria y Pesquera [SIAP, 2021]). Wilt caused by Fusarium oxysporum f. sp. niveum (Fon) is the main fungal disease affecting watermelon crops worldwide and can cause losses of up to 100 % when cultivars with resistance to the fungus are not used (Dau et al., 2009). This disease is characterized by presenting chlorosis, wilting of foliage, necrosis of vascular tissue and plant death (Callaghan et al., 2016; Fernández-Herrera, González-Soto, & Ramírez-Bustos, 2021). This pathogen survives in the soil for many years in the form of chlamydospores, which hinders watermelon cultivation in fields infested with this fungus (Kang, Demers, Jimenez-Gasco, & Rep, 2014), or it can infect seeds latently, which can be an important inoculum source and contribute to severe disease outbreaks (Petkar & Ji, 2017).
Fon has four races (0, 1, 2 and 3), all of which are present in the United States, although in many other countries, including Mexico, the distribution and prevalence of these races is still unknown. The traditional method for the identification of Fon races is through inoculation of differential cultivars; however, this method is time-consuming, expensive and inaccurate (Everts & Himmelstein, 2015; Hudson et al., 2021), as several factors can have a significant effect on pathogen virulence (such as temperature, moisture, spore concentration, seedling age, etc.). In addition, certain differential cultivars such as Calhoun Gray and PI-296341-FR are not commercially available, although Calhoun Gray can be substituted for Dixielee (Kleczewski & Egel, 2011; Zhou & Everts, 2003).
In contrast, molecular identification by PCR has advantages over traditional diagnostic methods in that it is faster, more sensitive and more reliable. Recently, Hudson et al. (2021) developed a primer pair that can differentiate Fon race 3 from races 1 and 2, and by using two other oligonucleotide pairs (Lin et al., 2010; Niu et al., 2016) races 1, 2 and 3 can be differentiated. However, that study did not include race 0 isolates, so it has yet to be identified based on inoculation of differential plants. A rapid and reliable identification of pathogens, or correct disease diagnosis, is an essential step in integrated disease management of agricultural crops (Lin et al., 2010).
In recent years, mainly during the spring-summer growing seasons, plants with symptoms similar to those described for F. oxysporum have been observed in commercial watermelon plantations in Hermosillo, Sonora, Mexico. Therefore, the aim of this research was to identify the race of Fusarium oxysporum causing wilt, vascular necrosis and death of watermelon plants in Sonora, Mexico, by using specific primers and inoculation of the isolates into commercially available differential watermelon cultivars.
Materials and methods
Sample collection and isolation
Watermelon plants with symptoms of wilt and vascular necrosis (Figure 1A, 1B) were collected from an agricultural field located in the municipality of Hermosillo, Sonora, Mexico (29° 01’ 39’’ NL and 111° 28’ 02’’ WL). Stems from three symptomatic plants of the triploid Joy Ride (with partial resistance to race 1, and susceptible to Fon race 2 and 3) were washed with copious amounts of water, then internal ~1 cm sections of the stems were cut, superficially disinfected with 1% sodium hypochlorite for 1 min, rinsed three times with sterile distilled water, and placed on sterile blotting paper. Tissue fragments were seeded in PDA medium (Difco®) and incubated at 28 °C for 5 days. Pure cultures of the isolates were obtained from a single spore. For morphological identification, tissue mounts from 10-day-old cultures were made. Three representative isolates (one isolate per plant) named UESFON01, UESFON02 and UESFON03 were inoculated into watermelon plants, with one true leaf, of the cultivar Sugar Baby, which is considered universally susceptible, since it lacks resistance genes to all known Fon races.
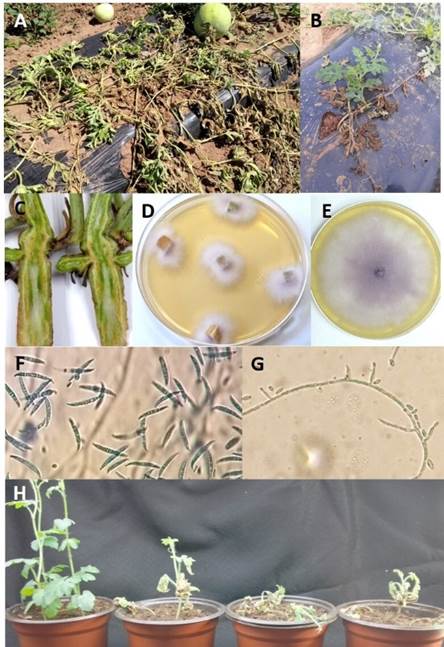
Figure 1 Symptoms and morphology of F. o f. sp. niveum isolated from watermelon plants with symptoms of wilt and vascular necrosis: A) wilt under field conditions, B) wilt on one side of the plant only, C) vascular necrosis on stem, D) isolation on PDA medium, 72 h after incubation (ai), E) growth on PDA at 8 days ai, F) macroconidia, G) short phialides and H) pathogenicity tests (from left to right: control, isolates UESFON01, UESFON02 and UESFON03).
Inoculation was performed on 15 watermelon seedlings per isolate, by pipetting 5 mL of a conidial suspension (1 x 106 spores∙mL-1) into the substrate around the plant. Control plants were only treated with sterile water (Latin & Snell, 1986). After the treatments, plants were kept in a growth chamber at 30 ± 2 °C.
Molecular identification of races of F. oxysporum f. sp. niveum
The 3 % CTAB method (Zhang, Uyemoto, & Kirkpatrick, 1998), with some modifications, was used for DNA extraction. A piece of mycelium was obtained from each of the three monoconidial fungal isolates, transferred to a 1.5 mL tube and macerated with 800 µL of 3 % CTAB extraction buffer, then incubated at 60 °C for 30 min. Samples were extracted with chloroform-isoamyl alcohol (24:1), 15 µL RNase A (1 µg∙µL-1) was added and then the samples were incubated at 37 °C for 10 min. The aqueous DNA layer was precipitated with 600 µL of isopropanol at -20 °C. The DNA pellet was washed with 70 % ethanol and dried at room temperature, then resuspended in 30 µL of nuclease-free water (Invitrogen) and stored at -20 °C for further analysis. DNA quality and integrity were assessed by electrophoresis on 1% agarose gels and by spectrophotometry (A260/A280) with a Nanodrop (ND-1000-UV-Vis, Nanodrop Technologies, USA).
DNA extracted from the fungal isolates was analyzed by PCR with three different primer pairs, which allow differentiating Fon races 1, 2 and 3 (Table 1). The first Fon-1/Fon-2 primer pair, specific for Fon, amplifies a 174 bp region (Lin et al., 2010). The second FONSIX6F/FONSIX6R pair amplifies a 453 bp fragment and allows differentiating Fon race 2 isolates from race 1 and 3 isolates, based on the absence of the SIX6 gene (Niu et al., 2016). Finally, FNR3-F and FNR3-R amplify 511 bp and allow differentiating Fon race 3 isolates from race 1 and 2 isolates (Hudson et al., 2021). PCR reaction mixtures were performed in a 25 µL volume, which contained 1 µL of DNA, 1X of buffer, 2 mM of MgCl2, 0.2 µM of each primer pair, 0.2 mM of each triphosphate nucleotide (dNTP) and 1 unit of Taq polymerase (Invitrogen Life Technologies, Brazil). Reactions were carried out in an automatic thermal cycler (C1000 Thermal Cycler, BioRad, USA).
Table 1 Characteristics of the oligonucleotides used in this study.
| Name | Primer (5’ - 3’) | Size (bp) | Specificity* | Reference |
|---|---|---|---|---|
| Fon-1 | CGATTAGCGAAGACATTCACAAGACT | 174 | Fon | Lin et al. (2010 |
| Fon-2 | ACGGTCAAGAAGATGCAGGGTAAAGGT | |||
| FONSIX6F | CGCTCTTATCGCATCAATCT | 453 | Fon2 | Niu et al. (2016 |
| FONSIX6R | GGGTTGACTGAGGTCGTGGT | |||
| FNR3-F | CGGCTTTCCTCTGTCAGATAGT | 511 | Fon3 | Hudson et al. (2021 |
| FNR3-R | TAGTGAGGTCCATGCCACGAA |
*Fon = Fusarium oxysporum f. sp. niveum; Fon2 = Fon race 2; Fon3 = Fon race 3.
The amplification conditions are described below. The Fon-1/Fon-2 primers had an initial denaturation at 94 °C for 90 s, followed by 30 cycles of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 60 s, with a final extension of 72 °C for 10 min. The FONSIX6F/FONSIX6R primers had one cycle amplification at 95 °C for 3 min, 30 successive cycles of 94 °C for 1 min, 66 °C for 1 min, and 72 °C for 1 min, with a final extension cycle at 72 °C for 10 min (Branham, Levi, & Wechter, 2019). The FNR3-F/FNR3-R primers underwent initial denaturation at 95 °C for 3 min, followed by 35 cycles at 95 °C for 30 s, 63 °C for 30 s, and 72 °C for 40 s, with a final elongation at 72 °C for 6 min. PCR products were analyzed by electrophoresis on 1% agarose gels stained with ethidium bromide (2 mg∙µL-1) and visualized in a UV transilluminator (Gel Doc XR+ Gel Documentation System, Bio-Rad, USA). Amplicons from the Fon-1/Fon-2 primers were purified and sequenced in both directions using the respective primer pair separately. For sequencing, an automated sequencer (3500 and 3130 Series Genetic Analyzer, Thermo Fisher Scientific, USA) was used.
Inoculation in differential watermelon plants
The three Fon isolates were inoculated into the differential watermelon cultivars Sugar Baby (without resistance genes to Fon 0, 1, 2 and 3), Charleston Gray (resistant to Fon 0, susceptible to Fon 1, 2 and 3) and Dixielee (resistant to Fon 0 and 1, susceptible to Fon 2 and 3), in order to avoid confusing race 0 with race 1. For this, seeds of the three differential cultivars were sown in sterilized Peat Moss, in germination trays with 50 cavities with a volume of 62.5 cm3 per cavity. After 15 days, 30 plants with a true leaf were inoculated into the substrate around the plant with 5 mL of a conidial suspension (1 x 106 spores∙mL-1). Control plants were only treated with sterile water (Latin & Snell, 1986; Pal, Rao, Thontadarya, Chandran, & Sriram, 2020). Spores of each fungal isolate were obtained by adding 30 mL of sterile distilled water to PDA dishes with the fungus grown for 10 days. The mycelium was scraped superficially and filtered through sterile gauze to remove mycelial fragments and adjusted to a concentration of 1 x 106 spores∙mL-1. The assay was performed twice under similar experimental conditions.
Watermelon cultivars were considered susceptible if they showed ≥33 % of plants with wilt symptoms or death, and resistant if they had <33 % wilted or dead plants after inoculation with the fungus (Zhou, Everts, & Bruton, 2010; Pal et al., 2020). Inoculated watermelon plants were evaluated individually in order to observe typical Fon symptoms (chlorosis, wilt, or plant death) up to 28 days after inoculation.
Results and discussion
Isolation and morphological identification
Only Fusarium oxysporum was isolated from watermelon plants with symptoms of wilt and vascular necrosis. Plants collected in the field showed chlorosis, generalized wilting (Figure 1A) or wilting on only one side of the plant (Figure 1B). Also, necrosis and discoloration of the vascular tissue of the stem was observed, which can be easily distinguished with a longitudinal or transversal cut in the lower part of the stem (Figure 1C). Three representative isolates of Fusarium oxysporum (with identification keys UESFON01, UESFON02 and UESFON03) from different symptomatic plants were selected for identification at race level.
Fusarium isolates showed the following morphological characteristics: pink to purple mycelium (Figures 1D and 1E), macroconidia curved and pointed at the ends with three to five cells from 18. 76 to 41.9 µm long and 3.38 to 4.34 µm wide (Figure 1F), oval to ellipsoid-shaped unicellular microconidia from 6.1 to 12.8 µm long and 2.12 to 3.4 µm wide, and short monophialides (Figure 1G). Based on these morphological characteristics, isolates UESFON01, UESFON02 and UESFON03 were identified as Fusarium oxysporum (Leslie & Summerell, 2006). Likewise, all three isolates caused symptoms of chlorosis, wilt, and death when inoculated into watermelon cultivar Sugar Baby seedlings (Figure 1H).
Fusarium oxysporum is a complex of isolates capable of causing disease in a wide range of agriculturally important crops. F. oxysporum is characterized by a high degree of host specificity. Isolates that are pathogenic to a single host are grouped in a special form (Rana, Sahgal, & Johri, 2017). F. o. f. sp. niveum is the special form that is pathogenic to watermelon; however, there are more than 100 recognized special forms, and identification based on their spore morphology is not possible due to the great similarity between special forms and other Fusarium species. Therefore, molecular identification is an important alternative for a more accurate identification, as it has advantages over traditional diagnostic methods, since it is faster, more sensitive and more reliable (Lin et al., 2010; Martyn, 2014; Hudson et al., 2021).
Molecular identification with specific primers
PCR with the specific primers Fon-1/Fon-2 amplified a 174 bp fragment, which confirmed that isolates UESFON01, UESFON02 and UESFON03 belong to the special niveum form (Figure 2A). Likewise, amplification of 453 and 511 bp fragments was observed with primers FONSIX6F/FONSIX6R and FNR3-F/FNR3-R, respectively (Figure 2B and 2C).
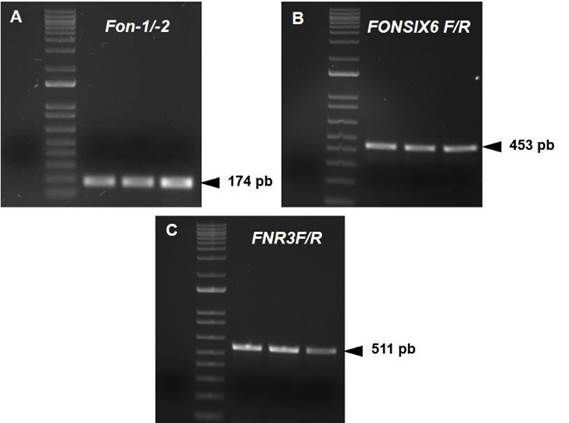
Figure 2 Amplification of Fon isolates with three primer pairs for identification of races 1, 2 and 3. A-C (from left to right): negative, molecular weight marker, UESFON01, UESFON02 and UESFON03.
The sequences of the amplicons obtained with the Fon-1/Fon-2 primers (OP060462, OP060463, OP060464) from the DNA of isolates UESFON01, UESFON02, and UESFON03 were deposited in the National Center for Biotechnology Information’s GenBank (NCBI; https://www.ncbi.nlm.nih.gov/). These sequences were compared using NCBI's BLAST program and showed 98 to 100 % similarity to Fusarium oxysporum f. sp. niveum (EU603504.1).
During infection in the host plant, Fusarium oxysporum f. sp. niveum secretes effector proteins into the xylem that can promote virulence or trigger resistance. SIX6 (secreted in xylem protein 6) is an avirulence gene that is present in races 0, 1 and 3, but not in race 2 (Niu et al., 2016). On the other hand, Hudson et al. (2021) performed a genomic analysis of Fon races 1, 2 and 3, and designed the FNR3-F/FNR3-R markers that amplify a 511 bp region, within a larger region (1,121 bp), on the pathogenicity chromosome that is absent in race 3, and present in races 1 and 2, making it possible to differentiate race 3 from races 1 and 2. Therefore, Hudson et al. (2021) point out that only race 1 isolates amplify with all three primer pairs, while the identification of Fon races 2 and 3 is determined by the absence of amplification when using the corresponding primer pair.
Based on the above, isolates UESFON01, UESFON02 and UESFON03 were identified as belonging to race 1. However, with these primers it is not possible to identify or determine race 0, since when race 0 isolates are amplified with these primers the results are variable (Keinath, DuBose, Katawczik, & Wechter, 2020; Hudson et al., 2021). Therefore, to avoid confusing the isolates with those of race 0, it is necessary to inoculate these into commercially available differential watermelon cultivars, which allow differentiating between Fon races 0 and 1 (Kleczewski & Egel, 2011; Zhou & Everts, 2003).
Inoculation into differential plants
The reaction to the disease (susceptible or resistant) of the differential cultivars, when inoculated with the three Fon isolates, was the same in the two trials conducted; therefore, only data from the first trial are shown in Table 2. In this trial, Sugar Baby plants (universally susceptible) showed 100 % mortality, Charleston Gray plants (race 0 resistant, race 1 susceptible) showed mortality ranges between 73.4 and 86.7 %, and Dixilee plants (race 0 and 1 resistant) showed between 6.7 and 16.7 % dead plants with the inoculation of the isolates. The low percentage of wilt or mortality of Dixilee cultivar plants inoculated with UESFON01, UESFON02 and UESFON03 confirm that these isolates belong to Fon race 1.
Table 2 Reaction of differential plants to inoculation with three isolates of Fusarium oxysporum f. sp. niveum 28 days after inoculation.
| Isolate/Differential | Number of inoculated plants | Mortality (%) | Reaction to the disease |
|---|---|---|---|
| UESFON01 | |||
| Sugar Baby | 30 | 100 | S |
| Charleston Gray | 30 | 76.7 | S |
| Dixilee | 30 | 16.7 | R |
| UESFON02 | |||
| Sugar Baby | 30 | 100 | S |
| Charleston Gray | 30 | 86.7 | S |
| Dixilee | 30 | 6.7 | R |
| UESFON03 | |||
| Sugar Baby | 30 | 100 | S |
| Charleston Gray | 30 | 73.4 | S |
| Dixilee | 30 | 13.3 | R |
Sugar Baby: universally susceptible; Charleston Gray: race 0 resistant; Dixilee: race 1 resistant. S = susceptible; R = resistant.
Vascular wilt of watermelon was first observed in 1890 in the southern United States, and since then this disease has been reported on all continents (except Antarctica), being the most important fungal disease of this crop worldwide (Martyn, 2014). There are four Fon races: 0, 1, 2 and 3 (Rahman et al., 2021), which are present in the United States and some other countries; however, in Mexico there is no information on the presence, distribution or prevalence of these races in the different watermelon-producing areas. There are many diploid watermelon cultivars with resistance to race 0 and race 1 isolates; however, increased production of triploid (seedless) watermelons, many of which lack high levels of resistance to race 1, has allowed a resurgence of Fusarium wilt (Everts, Egel, Langston, & Zhou, 2014; Everts & Himmelstein, 2015). For race 2, there are no cultivars with resistance to Fusarium, except PI296341- FR (Martyn & Netzer, 1991), while race 3, first reported in Maryland, is highly virulent to all watermelon cultivars, including PI296341- FR (Zhou et al., 2010).
Fon race 0 is of little economic importance because most cultivated cultivars are resistant to it (Everts & Himmelstein, 2015), whereas Fon race 1 is considered the most dominant race in watermelon-producing areas of the world (Rahman et al., 2021), such as India (Pal et al., 2020), Turkey (Kurt et al., 2008) and China (Zhong et al., 2022). However, race 2 is currently an emerging problem and has been reported to be more prevalent than race 1 in several regions (Zhou & Everts, 2003; Keinath et al., 2020), such as Israel (Netzer, 1976), Spain (Gonzalez-Torres, Meléro-Vara, Gómez-Vázquez, & Jiménez-Díaz, 1993) and China (Duan et al., 2007). The increase in race 2 populations is favored by the use of cultivars with resistance to races 0 and 1 (Hopkins, Lobinske, & Larkin, 1992; Zhou & Everts, 2007).
In Mexico, despite the great importance of watermelon cultivation, there is no information on the Fon races associated with wilt or plant death in watermelon cultivation. In this study, based on the symptoms described, morphological characteristics, amplification with the specific primers Fon-1/Fon-2, FONSIX6F/FONSIX6R and FNR3-F/FNR3-R, and inoculation of three differential watermelon cultivars, it can be concluded that race 1 of Fusarium oxysporum f. sp. niveum is part of the fungal complex that causes wilt and death of watermelon plants in Mexico.
Conclusion
Fusarium oxysporum f. sp. niveum race 1 was identified as a causal agent of wilt and plant death in the watermelon crop in Hermosillo, Sonora, Mexico. This is the first report of Fon race 1 in watermelon in our country, and it will help to better understand the fungal diseases that threaten this crop. Likewise, it will allow the development of integrated control strategies to improve the production of this crop.











 texto em
texto em 


