Introduction
The Fusarium wilt produced by the fungus Fusarium oxysporum f. sp. cubense (FOC1; discovered in 1874), caused great devastation in the banana sector during the first half of the 20th century, due to the sensitivity of the Gros Michel clone, which led to its replacement with the Grand Nain clone beginning in the 1950s and which remains as the predominant commercial variety worldwide (Wang et al., 2012). In the 1990s, a new fungus strain was identified in Southeast Asia called Fusarium oxysporum f. sp. cubense (FOC R4T), which affects the Cavendish cultivar, cooking bananas, edible diploids, and plantains. Due to the increasing dispersion of FOC, one of the bases for integrated disease management is knowledge of the niche occupied by the new pathogen and how this affects the host's living conditions. Therefore, it is vital to know the soil microbiota and the phytobiome, particularly the species associated with the roots. In this context, it is important to analyze the communities of microorganisms that can interact with FOC under different farming systems (Shi et al., 2013).
Soil is an ecosystem with a great variety of microbial populations, which directly influence its activity and plant growth. Microorganisms are the first to react to physical and chemical changes in the environment; therefore, it is of vital importance to analyze their behavior and interactions. This enables establishing conditions where FOC can be suppressed from the rhizosphere of a banana crop pathosystem (Gryta, Frąc, & Oszust, 2014). Community physiological profiling has proven to be very effective in determining spatial and temporal changes in soil microorganism communities. The evaluation of data generated from miniaturized metabolic tests include average well color development (AWCD), the richness index and Shannon diversity (Frąc, Oszust, & Lipiec, 2012). AWCD is a general indicator of the metabolic activity of the microbial community and can be subdivided among the groups that have a similar chemical nature, allowing an evaluation of the microbial community’s potential to degrade different carbon sources (Kenarova, Radeva, Traykov, & Boteva, 2014). Molecular identifications of soil indicators that are present in soil communities and their comparison is an adequate way to find differences in agroecosystem functions.
Soil organisms are an integral component of ecosystems, but their activities receive little recognition in management strategies. Land-use intensity is constantly increasing on a global scale with adverse effects on soil ecosystems (Stavi & Lal, 2015). One-quarter of all soils worldwide face degradation and an increasing number of studies show that intensive land use threatens soil biodiversity. To enhance soil biodiversity, specific changes in soil community composition can complement each other to increase ecosystem sustainability (Tsiafouli et al., 2015) and also improve soil health, in terms of promoting soil resiliency and being more suppressive against some pathogens. In general, some studies demonstraded that soil microorganisms are involved in the nitrogen and carbon cycles and also enhance plant nutrition (Bommarco, Kleijn, & Potts, 2013).
In natural ecosystems, wild plants are dependent on the activity and function of their rhizosphere communities for defenses against soil pests and diseases (Huang et al., 2013). Over evolutionary time, plants developed intimate relationships with beneficial soil microorganisms, taking advantage of their ability to inhibit plant pathogens. Currently, there are a lot of molecular tools that allow scientists to improve our knowledge of the soil microbiome and the functions of the microorganisms associated with plants. Engineering plant-soil biota interactions (through soil inoculation), genome editing, metagenome sequencing and plant trait selection, offers a promising avenue to rapidly face the challenges in both natural and agricultural systems in the future (Mariotte et al., 2018).
A current challenge is to go beyond predictive understanding of genome function based on the genome and metagenome to understanding the actual functions carried out by the soil microbiome in situ. Some scientists discovered that to control or manage FOC R4T, it is necessary to apply other more efficient and effective management strategies (such as biological control), with the use of arbuscular mycorrhizal fungi (AMF) and plant growth promoting rhizobacteria (PGPR) possibly being needed to overcome the disease (Siamak & Zheng, 2018). Other authors suggest that the successful colonization of host rhizospheres is very important for effective control of soil-borne pathogens, and they have demonstrated that the combined application of endophytic and rhizospheric bacteria strains results in effective suppression of FOC (Ploetz, 2006; Thangavelu & Gopi, 2015).
The future of FOC control will be based on complete knowledge of the natural microbe soil which has a very important role in pathogen control and also improves plant growth, soil health and soil resiliency to all climate change. The microbial populations and community made the soil more disease-suppressive and beneficial to soil nutrient cycling and plant growth (Xinqi et al., 2015). Therefore, this study aimed to establish differences in microbial communities associated with crops of Gros Michel banana clone and biochemical profiles of these communities, in farms under different agronomic conditions, to improve our understanding of factors that can contribute to the effective control of banana fusariosis.
Materials and methods
Farm selection and location
Seven farms producing Gros Michel banana clone were selected in Turrialba, Costa Rica, where three different agricultural conditions were evaluated: organic (9° 53’ 57.63” NL - 83° 41’ 41.62” WL and 9° 52’ 41.7” NL - 83° 42’ 23.18” WL), intermediate conventional (9° 56’ 43.75” NL - 83° 39’ 44.4” WL and 9° 56’ 3.28” NL - 83° 37’ 51.07” WL) and abandoned (9° 56’ 3.61” NL - 83° 37’ 47.8” WL and 9° 56’ 6.84” NL - 83° 37’ 47.42” WL), with high or low fusariosis severity in each condition. A forest ( 9° 56’ 46.22” NL - 83° 41’ 25.07” WL) was used as the control. Organic farms were certified as organic according to international organisms, and following standards. Also, the farms must have had at least 2-3 years of applying the concepts of organic agriculture. Conventional farms apply conventional agronomic managment, in which the agriculturist uses synthetic chemicals in order to control diseases and to fertilize. Abandoned farms were sown with banana in the past, but have been been abandoned for at least 5 years. In this case the banana plants cotinued to grow without any human intervention.
Biochemical profile determination of microbial communities associated with FOC TR1
EcoPlates (Biolog®, USA) were used to measure microbial functional diversity. A similar protocol to that described by Velkeneers (2013) was followed, where 5 g of soil were resuspended in 50 mL of phosphate buffer (0.05 M, K2HPO4, pH 7), inoculated with 150 µL of dilution per well and kept at 26 °C. Readings were taken every 18 h. The AWCD (average well color development) was calculated using the formula:
where OD i is the optical density and 31 is the number of carbon substrates present in the EcoPlates. The richness index (RI) was obtained by counting the number of positive reactions (standardized OD higher than 0.25) with the following formula:
where R is the number of total carbon substrates (31). The Shannon-Weaver (SW) index was calculated using the formula:
where ODi(31) is the total carbon substrates in the EcoPlates and ODi(96) is the total number of wells (three replicates of each substrate, three negative water controls) of the EcoPlates.
By grouping by substrates the AWCD values of each farm at 54 h, a principal component analysis (PCA) was conducted to determine the groups of substrates that are mostly metabolized. A similar analysis was performed by Ge, Du, Gao, and Qiu (2018).
Microbial indicators
For testing the presence of different important groups in the soil community, seven microorganisms were selected as indicators (Table 1). These microorganisms were selected according to the literature where the importance of some specific microorganism groups as indicators of soil health and the principal pathogens of banana plants are described (Xinqi et al., 2015). Primers were designed for indicator species using Primer 3Plus bioinformatics software. The microorganisms present in the soil were determined through PCR (polymerase chain reaction). The Power Soil® DNA Isolation (MoBio Laboratories Inc, UK) extraction kit was used. The integrity of the extracted DNA was corroborated by electrophoresis in agarose gel. Sanger sequencing of the amplified DNA bands was made to determine the specific identity of the microorganisms. Sequence comparisons were made with the NCBI (National Center for Biotechnology Information) database.
Table 1 Microorganism groups selected for molecular identification.
| Microorganisms | Function | Primer sequence | Tm (°C) |
|---|---|---|---|
| Azospirillum sp. | Phosphorus solubilizer | F: TCGGCCTTAGTGGCGCACGGGT R: GGCCGTGTCTCAGTCCCAGTGTGGC | 67.2 |
| Rhizobium sp. / Azotobacter tropicalis | Nitrogen fixative | F: TGAGGTGCGAAAGCGTGGGGAGCAA R: TGTGCGGGCCCCCGTCAATTCCTTT / F: AAAGCCTGATCCAGCCATGCCGCGT R: TGAACCACCTACGCGCTTACGCCCA | 66.7 / 66.2 |
| Nostoc sp. | Photosynthesizers | F: ATTGGGCGTAAAGCGTCCGCAGGT R: AGCGCCTTCGCCACTGGTGTTCTT | 65.3 |
| Streptomyces sp. | Producer antibiotic substances | F: GCACAAGCAGCGGAGCATGTGGCTT R: AAGGGTTGCGCTCGTTGCGGGACTT | 66.3 |
| Ralstonia sygygii | Pathogen indicator | F: AAGTCCCGCAACGAGCGCAACCCTT R: ATTAGCTCCACCTCGCGGCTTGGCA | 67.1 |
| Bacillus subtilis | Cellulose degrader | F: ACCCGCGGCGCATTAGCTAGTTGGT R: ACGCGGCGTTGCTCCGTCAGACTTT | 66.6 |
Results
Biophysical and environmental diagnosis
Table 2 summarizes the predominant biophysical characteristics in each of the sampled farms. Disease severity was classified as high or moderate. Low severity was when less than the 50 % of the total plants in the farm presented moderate symtoms. High severity was assigned when more than 90 % of the total plants in the farm were diseased, presenting all the symptoms of Fusarium wilt in the last stage.
Table 2 Physical characteristics of each farms sampled.
| Farm | Fusariosis severity | Altitude (m) | Temperature (°C) | Soil type | Relative humidity (%) |
|---|---|---|---|---|---|
| 1 - Organic | Low | 1,000.69 | 25 | Andisol | 77 |
| 2 - Organic | High | 1,188 | 25 | Andisol | 77 |
| 3 - Conventional | High | 763.6 | 29 | Andisol | 72 |
| 4 - Conventional | Low | 465.19 | 22 | Inceptisol | 82 |
| 5 - Abandoned | Low | 524.01 | 20 | Inceptisol | 80 |
| 6 - Abandoned | High | 427.9 | 22 | Inceptisol | 82 |
| 7 - Forest | - | 1211 | 28 | Andisol | 70 |
Biochemical profile of the microbial communities associated with FOC R1
AWCD values were transformed using log10 and showed changes in microbial abundance and metabolic activity (Figure 1). An increment in absorbance was observed as the incubation time progressed. The abandoned farms (5 and 6) showed the fastest reaction and farm7 the slowest.
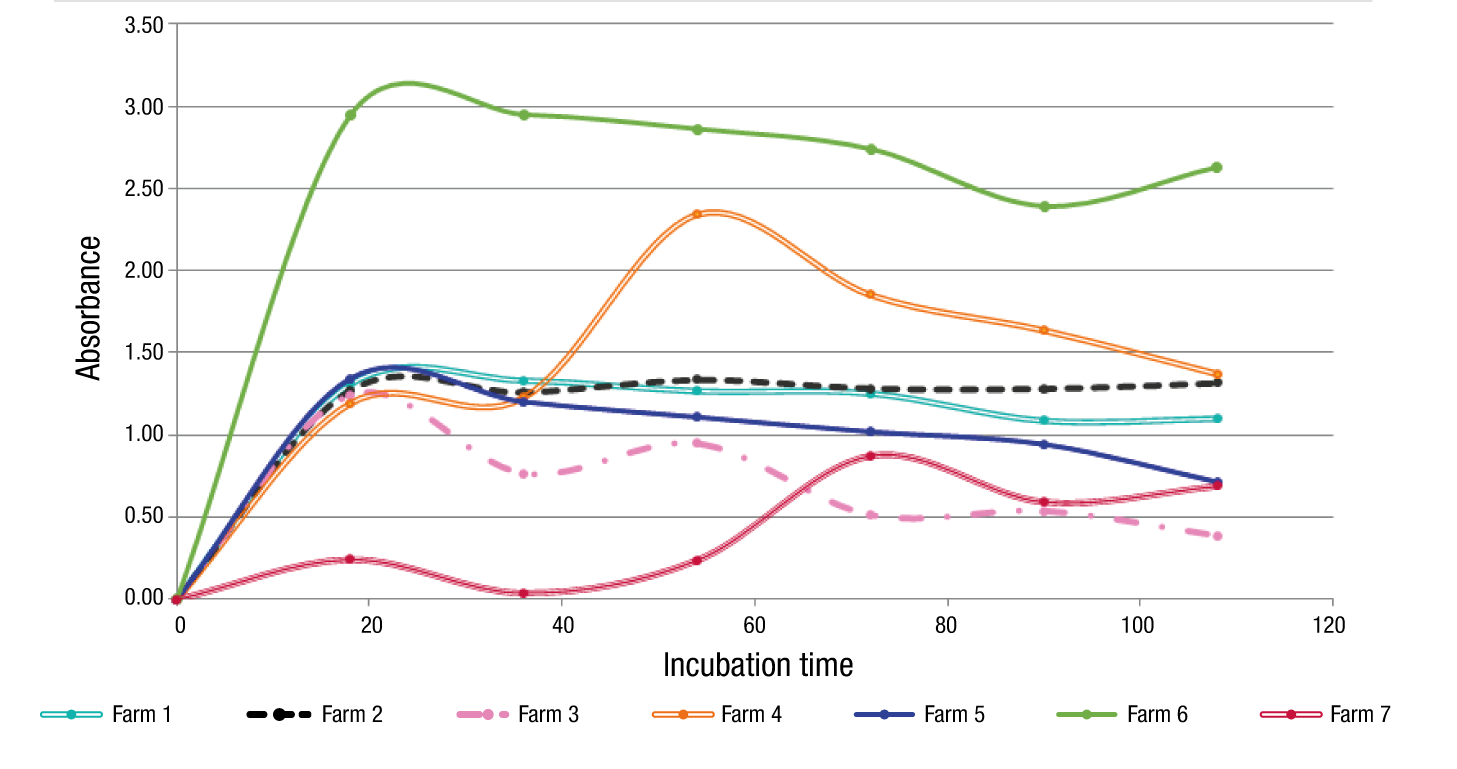
Figure 1 Average well color development (AWCD) of the seven tested farms until 108 h of incubation. Farms 1 y 2 = organic; Farms 3 y 4 = intermediate conventional agronomic management; Farms 5 y 6 = abandoned; Farm 7 = forest (negative control).
Farm 6 had the greatest absorbance values from 20 to 108 h, which means a high metabolic response to the substrates. Farms 1, 2 and 5 had similar behavior in their profiles throughout the experiment. Farms 3 and 7 showed a profile under 1 in the absorbance value.
The total number of carbon sources metabolized by soil microorganisms relative to the total number of substrates shows the specific richness (Figure 2). It was observed that the largest number of metabolized substrates was obtained at 108 h, farm 6 (abandoned) being the one that presented the highest value. In terms of diversity, the results show that farm 2 had the largest functional diversity at 108 h of incubation (Figure 3).
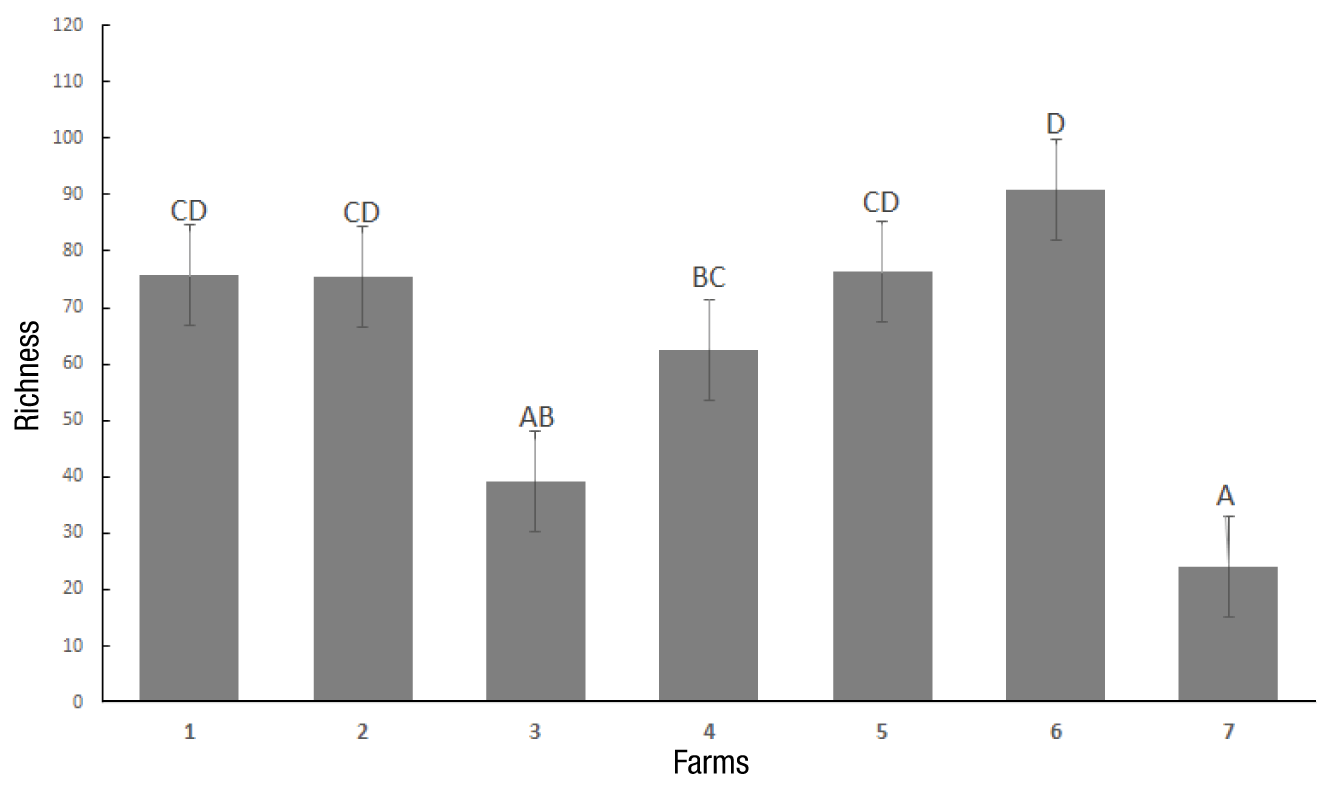
Figure 2 Richness index of the microbial community of the seven tested farms until 108 h of incubation in Ecoplates at 26 °C. Farms 1 y 2 = organic; Farms 3 y 4 = intermediate conventional agronomic management; Farms 5 y 6 = abandoned; Farm 7 = forest (negative control). zSame letter within each bar do not differ statistically (Kruskal-Wallis, P ≤ 0.001).
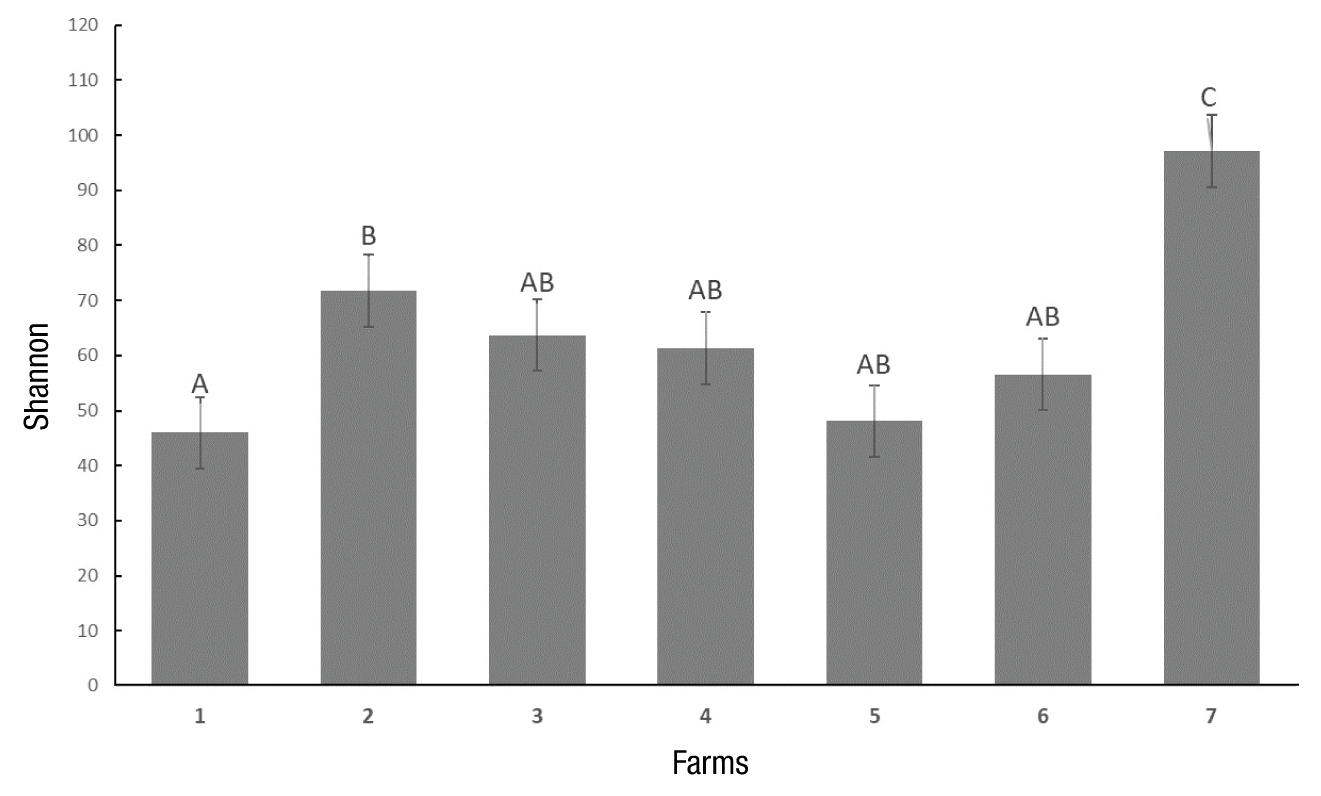
Figure 3 Shannon-Weaver index of the microbial community of the seven tested farms until 108 hours of incubation in Ecoplates at 26 °C. Farms 1 y 2 = organic; Farms 3 y 4 = intermediate conventional agronomic management; Farms 5 y 6 = abandoned; Farm 7 = forest (negative control). zSame letter within each bar do not differ statistically (Kruskal-Wallis, P ≤ 0.004).
A PCA was conducted. The first two components encompassed 88.8 % of the variance (Figure 4). PC1 shows an important positive correlation with amino acids, polymers, and carboxylic acids. At the same time, there is a strong negative correlation with amines. PC2 has a high and positive correlation with carbohydrates, moderately important with amines and a strong negative correlation with phenols. Farms 3 and 7 are positively influenced by PC1, while farms 2 and 3 are positively influenced by PC2. In the case of farms 1, 6, 5 and 4, they are very close to value 0 or graph axis. Farm 4 also appears to be more associated with phenolic compounds (Figure 4).
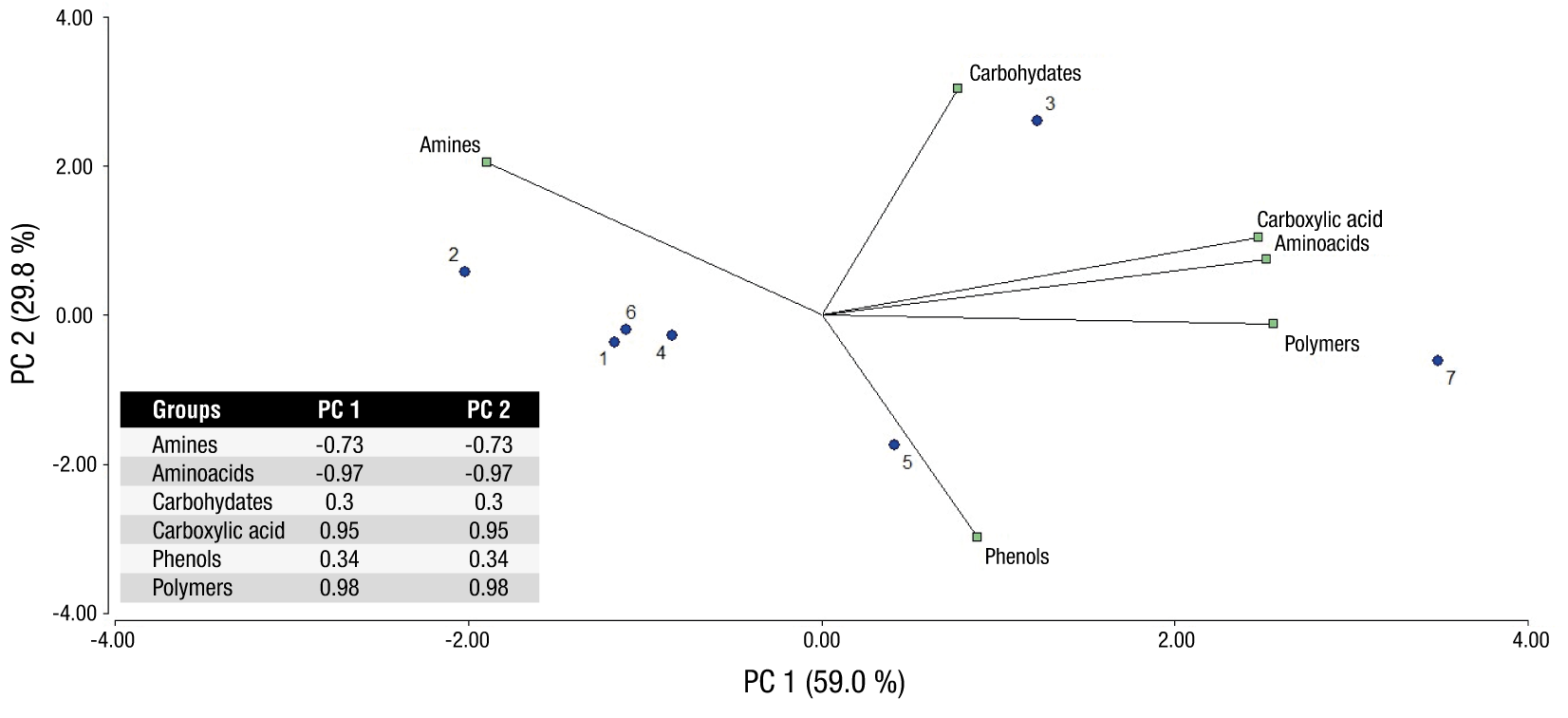
Figure 4 Bi-plot graph of the principal component analysis obtained from the analysis of average well color development (AWCD) at 54 h of incubation in EcoPlates. Farms 1 y 2 = organic; Farms 3 y 4 = intermediate conventional agronomic management; Farms 5 y 6 = abandoned; Farm 7 = forest (negative control).
Microbial indicators
All designed primers were evaluated with seven sampled farms to corroborate the presence or absence of different microorganisms. Microorganisms identified with an identity percentage above 95 % are shown in Table 3. It was observed that among farms with the same conditions, different species of bacteria were found; although Bacillus was identified on all farms. The presence of Ralstonia in several of the analyzed soils indicates that they have ideal conditions for the development of root pathogens. In general, the soils are very diverse in terms of the indicator microorganisms that were found.
Table 3 Microorganisms identified on each farm.
| Farm | Microorganisms found | Identity (%) | E-value |
|---|---|---|---|
| 1 | Nostoc | 94 | 2e-19 |
| Bacillus sp. | 99 | 6e-64 | |
| 2 | Ralstonia sp. | 97 | 6e-62 |
| Rhizobium | 97 | 4e-08 | |
| Bacillus subtillis | 99 | 2e-51 | |
| 3 | Ralstonia sp. | 94 | 1e-46 |
| Streptomyces | 95 | 4e-08 | |
| Bacillus subtillis | 100 | 9e-63 | |
| 4 | Azospirillum | 95 | 3e-21 |
| Nostoc sp. | 100 | 5e-07 | |
| Bacillus sp. | 100 | 6e-60 | |
| 5 | Ralstonia sp. | 95 | 1e-50 |
| Bacillus subtillis | 100 | 7e-56 | |
| 6 | Nostoc sp. | 100 | 3e-07 |
| Bacillus sp | 100 | 1e-56 | |
| 7 | Ralstonia sp. | 98 | 1e-63 |
| Bacillus subtillis | 100 | 4e-66 |
Chemical indicators
As can be seen in Table 4, farms 3 and 7 have acidity problems, while their percentages of organic matter are quite elevated. All effective cation exchange capacity (ECEC) and micronutrient values were found within the established range for moderately fertile soils (Estrada-Herrera et al., 2017).
Table 4 Chemical analysis of the seven types of soil sampled.
| Farm | pH | Acidity | Ca | Mg | K | ECEC | AS (%) | (Ca+Mg)/K | P | Fe | Cu | Zn | Mn | B | OM (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.32 | 1.23 | 4.39 | 0.91 | 0.16 | 12.63 | 9.74 | 33.13 | 6 | 150 | 6.1 | 1.3 | 15 | 0.15 | 16.7 |
| 2 | 5.62 | 0.30 | 19.90 | 4.77 | 0.44 | 28.27 | 1.06 | 56.07 | 26 | 271 | 3.3 | 6.4 | 88 | 0.31 | 11.2 |
| 3 | 4.26 | 13.00 | 5.44 | 2.29 | 0.27 | 25.93 | 50.13 | 28.63 | 10 | 247 | 5.2 | 2.4 | 104 | 0.14 | 8.6 |
| 4 | 5.88 | 0.22 | 20.70 | 4.92 | 0.70 | 29.94 | 0.73 | 36.60 | 26 | 229 | 4.1 | 7.9 | 74 | 0.38 | 10.5 |
| 5 | 6.02 | 0.14 | 16.00 | 3.81 | 0.60 | 28.35 | 0.49 | 33.02 | 41 | 291 | 8.4 | 4.3 | 99 | 0.29 | 7.2 |
| 6 | 5.74 | 0.24 | 13.30 | 3.83 | 0.49 | 19.57 | 1.23 | 34.96 | 26 | 164 | 2.2 | 1.2 | 49 | 0.28 | 11.4 |
| 7 | 4.68 | 3.08 | 1.21 | 0.47 | 0.26 | 7.66 | 40.21 | 6.46 | 13 | 222 | 2.9 | 2.1 | 16 | 0.19 | 35.7 |
ECEC = effective cation exchange capacity; AS = acidity saturation ; OM = organic matter.
Discussion
Metabolic analysis of microbial communities involves the characterization of biochemical patterns that enables establishing the consumption of carbon sources in a specific time. In general terms, the diversity-function relationship can be determined from the microbial community present (Arteaga-Garibay, Gómez-Estrada, Martínez-Peña, Cadena-Zamudio, & Avendaño-Arrazate, 2016).
The obtained metabolic profile showed that abandonment and agronomic intervention are two processes that make different microbial communities. In this case, the highest levels of AWCD were present in the conventional management (4) and abandoned (6) farms. However, in terms of richness and diversity, the organic farms (1 and 2) remained among the highest indices along with the abandoned (6) and control (7) farms.
The AWCD profile from the two organic fields is similar, but they are very different from the other systems (conventional and abandoned systems). Some authors such as Liu, Tu, Hu, Gumpertz, and Beagle-Ristaino (2006) indicate that farms under organic management are the ones that show the largest differences relative to other types of management, which agrees with our observations, where farm 4 had higher AWCD values compared to the homogeneous behavior of the two organic farms. Nonetheless, this can be partly due to the extent of intervention on each farm.
Liu et al. (2006) found significant differences in the functional richness of microbial communities, obtaining higher richness values in organic farms when performing comparative analyses between different types of agronomic management systems. In this study, high richness values were obtained in the organic farms (1 and 2), and statistical differences were observed with farm 3 (Figure 2).
The similarities found in the Shannon index suggests that management practices have not exerted a major influence on the functional structure of the soil microbial community (Lopes et al., 2010). Low Shannon-Weaver values suggest that members of different communities are very analogous with each other, which does not allow them to have high levels of functional diversity.
Deltour et al. (2017) analyzed the impact of FOC at different locations on a single farm managed as an agroforestry system based on agroecological principles (without the use of synthetic fertilizers), and they found that the area with the highest levels of FOC suppression was the one managed on the basis of a total agroforestry management system since 1995; it has some specific characteristics, such as higher soil clay content, pH and electrical conductivity (EC) values. They also discovered that the areas with a higher degree of suppression had a higher diversity of other banana cultivars, a lower soil cover and a higher abundance of adult trees (Deltour et al., 2017).
It has been found that a mixture of at least two banana varieties, with different levels of Fusarium wilt resistance is associated with a higher degree of disease suppression (Karangwa, Blomme, Beedd, Niyongere, & Viljoen, 2016). As bananas differ in their biochemical root composition and root exudation, they might be colonized by different microbial communities and thus modulate the soil pathogens (Wuyts et al., 2007).
Shen et al. (2019) studied the use of synthetic fertilizers and chemical pesticides (such as ammonia) in conventional agronomic management and found that fumigated soils exhibited a lower abundance of fungal microbes and FOC; however, there was a non-significant difference in the bacterial community in the two cases: fumigated soils and non-fumigated soils. Also, the Shannon diversity showed no significant effect on the microbial community in both soils.
Biofertilizer application after soil fumigation modified the disturbed soil microbial community induced by banana mono-cropping, leading to a unique soil microbial community. This is an indicator that in some cases organic amendments (like biofertilizers) applied to soil after fumigation, may be beneficial for diversification of microbial community, which was reported to result in reduced FOC in the soil (Shen et al., 2019; Wang et al., 2012).
The PCA showed a high positive correlation of carboxylic acids, polymers and amino acids found in PC1 denote the importance of these substances in soil microbial dynamics. These compounds are part of organic matter, which is constituted by animal and plant residuals in different degrees of decomposition; most of these compounds are carbohydrates, cellulose, lignin, proteins, amino acids, and waxes. All these substances are primary sources of nutrients for macro and microorganisms (Schmidt et al., 2011). Carboxylic acids can also be exuded by plants, through the roots (this can occur due to stress caused by lack of nutrients); with these exudates, in turn, providing food and energy for microorganisms in the soil (López-Reyes et al., 2012). In soil renewal processes, mainly the most complex polymers, carboxylic acids, and amino acids are metabolized (Food and Agriculture Organization of the United Nations [FAO], 2006).
Regarding the amines, in PC1 they are not consumed but in PC2 they are the second factor of consumption; this makes the points very close to the central axis in the graph, which may suggest that farms located close to it do not show a clear predilection for a given substrate. The two organic farms (1 and 2) did not show significant statistical differences between them, both in the physiological profile analysis as well as in one of the PC; therefore, it can be said that they have very homogeneous biochemical behavior.
Farm 7 (control) shows behavior highly affected by carboxylic acids, amino acids and polymers, therefore tending towards the positive values of PC1. Jangid et al. (2008) have detailed the chemical and biological differences with forests, which are frequently used as controls in agronomic studies of farms. Differences have been found in terms of carbon source usage by forest microorganisms relative to those of agricultural soils, and greater consumption of less energetic substrates such as carboxylic acids and amino acids has been observed. This is probably because forest cover has high concentrations of organic matter and without soil mechanical alteration lower pH values are exhibited, so the microorganisms present will have a greater preference for acid substrates (Brackin, Robinson, Lakshmanan, & Schmidt, 2013).
Concerning the diverse roles played by microorganisms in the soil, those groups with major importance for banana farms were chosen. Results show the presence of Bacillus bacteria at all sites, something previously reported by other researchers in banana soils (Fu, Ruan, Tao, Rong, & Shen 2016), where Bacillus spp. is catalogued as one of the most predominant species even under different types of agricultural management (organic and conventional). One of the main functions of Bacillus spp. is to degrade cellulose; however, some authors indicate that these bacteria can also play other vital roles in soils such as urea solubilization, nitrogen compound oxidation and antibiotic production (Chaparro, Sheflin, Manter, & Vivanco, 2011).
Other organisms were also searched for, such as Azospirillum spp. and Nostoc spp., with variable results in terms of their presence in different sampled areas. Results show that there is no evenness in microbial communities of sites under the same agronomic management, which agrees with other studies that describe microbiological differences in farms with organic management. Wessén, Hallin, and Philippot (2010) found that conducting different activities and applying different organic amendments to this crop modality cause distinct effects on soil, with metabolic activity being promoted in some cases but not in others. Therefore, under the same type of agronomic management, farming activities could make an important difference in terms of microbiome composition.
The results show that the farm with the greatest presence of beneficial indicator microorganisms is the one with conventional management (farm 4), in which the microorganisms Azospirillum spp, Nostoc spp., and Bacillus spp. stood out (Table 3), while the presence of Ralstonia spp. was not detected. The absence of this pathogen could be a signal of effective crop protection practices, and the presence of several beneficial indicators could indicate good soil management. Nostoc spp is characterized by being a photosynthesizer with the ability to fix atmospheric nitrogen; these characteristics allow it to be one of the first colonizers of decomposing matter, by providing oxygen to the environment through photosynthesis and thus allowing other aerobic bacteria to grow later (Obana, Miyamoto, Morita, Ohmori, & Inubushi, 2007). In addition, Azospirillum spp. is associated with phosphorus-rich compounds, solubilization and atmospheric nitrogen fixation and secretion of plant growth regulators (García-Orenes et al., 2010; Méndez-Gómez, Castro-Mercado, & García-Pineda, 2014).
Due to the high incidence of Bacillus found in the rhizosphere of all the sampled farms, it is considered a very important microorganism for soil health; it promotes plant growth and at the same time has an antibiotic effect against phytopathogens. Surprisingly, farm 7 (control) has the presence of Ralstonia and does not have the majority of cycle indicators.
The results obtained in soil chemical analysis show that all farms have an ECEC higher than the theoretical value established for a high fertility level (Estrada-Herrera et al., 2017). In zero tillage (abandoned) farms, high ECEC values can be seen. This characteristic favors the concentration of organic matter and nitrogen compounds; in addition, it prevents erosion, and decreases mineral leaching in the soil and general microbiological conditions (Sadzawka, 2011). The acidification that occurs in no-till systems does not necessarily mean an increase in interchangeable aluminum, because soil physical structure is not being altered, and the content of organic compounds increases, which are capable of catching Al ions, which decrease the toxicity of this compound for plants.
Conclusions
The metabolic profile, diversity and richness of each of the different farms were affected by the type of agronomic management used. But the composition of the communities in the rhizosphere of banana plants seems to be determined by a series of highly complex factors. Farm 4 (intermediate conventional) stood out, presenting the most favorable values in all factors analyzed, with all obtained values indicating that it is a metabolically very active soil under the presence of beneficial populations.
Differences in chemical and physical factors and the microbiome soil communities were observed under the different agronomic management systems. The sequencing analysis determined that genus Bacillus is the most predominant group in all farms, but differences were found regarding the presence of indicator microorganisms. There are important microorganism groups that reflect soil health and pathogen resistance (Baccillus, Azospirrillum, Nostoc, and Streptomyces). Microorganism communities established in the banana rhizosphere and the factors that affect them are some of the most unknown aspects of developing good farming practices.











 texto em
texto em 



