Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.34 n.2 Texcoco 2016
https://doi.org/10.18781/R.MEX.FIT.1601-2
Phytopathological notes
Identification and phylogenetic analysis of the leaf-galling nematode Orrina phyllobia affecting Solanum elaeagnifolium Cav. in Guanajuato, Mexico
1 Departamento de Producción Agrícola y Animal, Universidad Autónoma Metropolitana, Calzada del Hueso No. 1100, Col. Villa Quietud, Delegación Coyoacán, Distrito Federal, CP 0496, México
2 Catedrático CONACYT-Centro Nacional de Metrología (CENAM), Km 4.5 Carretera a Los Cués, El Marqués, Querétaro, C.P. 76246, México
3 Departamento de Parasitología, Universidad Autónoma Chapingo, Km. 38.5 Carretera México-Texcoco, Estado de México, CP 56230, México
4 Laboratorio de Microbiología, Universidad Autónoma Metropolitana, Calzada del Hueso No. 1100, Col. Villa Quietud, Delegación Coyoacán, Distrito Federal, CP 0496, México
The silver-leaf nightshade Solanum elaeagnifolium is an economic importance weed affected by natural enemies that would be used as biological control agent. The leaf-galling nematode Orrina phyllobia was detected parasitizing S. elaeagnifolium in San Luis de la Paz, Guanajuato. Morphotaxonomic and molecular analysis was performed on specimens extracted from foliar galls and distortioned leaves. The morphometric values were in agreement with those reported by the original species description except with de Man ratios: a and b. The Polymerase Chain Reaction amplification products of two molecular markers ITS1-5.8S-ITS2 and the D2-D3 expansion segments of 28S gene were 850 and 900 bp respectively. The homology by BLASTING of the mexican population genetic sequences together with those from NCBI database showed 99% similarity with Ditylenchus phyllobius (E=0.0). The bayesian inference of the ribosomal DNA marker ITS1-5.8S-ITS2 with sequences of genus and anguinid species uploaded in GenBank displayed a clear grouping affinity with Ditylenchus phyllobius. The combination of morphometric and genetic characters allowed to confirm the identity of O. phyllobia atacking S. elaeagnifolium in Mexico.
Key words: Biocontrol; nematodes; weeds; sequencing; phylogeny
El trompillo Solanum elaeagnifolium es una maleza de importancia económica afectada por enemigos naturales que podrían ser empleados como agentes de control biológico. Se detectó al nematodo agallador Orrina phyllobia parasitando S. elaeagnifolium en San Luis de la Paz, Guanajuato. Se realizó la identificación morfotaxonómica y molecular de especímenes extraídos de agallas y deformaciones foliares. Los valores de las características morfológicas y morfométricas concordaron con los rangos reportados en la descripción original de la especie a excepción de los índices de Man: a y b'. Los productos de la amplificación mediante la Reacción en Cadena de la Polimerasa de dos marcadores moleculares ITS1-5.8S-ITS2 y los segmentos de expansión D2-D3 del gen 28S fueron 850 y 900 pb respectivamente. La homología por BLASTING de secuencias genéticas de la población mexicana junto con secuencias de la base de datos del NCBI mostró un 99 % de identidad con Ditylenchus phyllobius (E=0.0). La inferencia bayesiana de la región ITS1-5.8S-ITS2 del ADN ribosomal con secuencias de géneros y especies de anguinidos depositados en el GenBank indicó una clara afinidad de agrupamiento con D. phyllobius. La combinación de caracteres morfométricos y genéticos permitió confirmar la identidad de Orrina phyllobia afectando S. elaeagnifolium en México.
Palabras clave: Biocontrol; nematodos; malezas; secuenciación; filogenia
The silver-leaf nightshade Solanum elaeagnifolium is a perennial plant from the Solanaceae family, endemic of northeast Mexico and southeast U.S.A. (Texas and Florida) (Cuda et al., 1988); it is considered an invasive plant and it is distributed in almost all over the world (EPPO, 2007). The area of Monterrey, Nuevo Leon, Mexico is considered the center of origin and evolution of this plant, based on the huge diversity, distribution, and frequency of natural enemies it displays (Wapshere, 1988). It is a weed of economic importance in several crops, particularly in intensive agriculture in northern Mexico. It is also considered a ruderal species as a result of the habitats altered by human activity, and can be commonly found alongside roads, abandoned agricultural fields, and even in urban areas (Vibrans, 2009). Due to its capacity as pests and diseases reservoir, cultural management measures have been taken as part of the plant health regulation to restrict its establishment and spreading of insects and other phytophagous (DOF, 1997). Among the organisms associated to S. elaeagnifolium and that could be used as possible biological control agents, different orders and families of insects stand out, along with gall mites, and nematodes. The leaf-galling nematode Orrina phyllobia (Thorne, 1934) Brzeski, 1981, was initially reported as Ditylenchus phyllobius sin. Nothanguina phyllobia (Orr.) and is distributed in southern U.S.A and southern India, affecting this weed (Brzeki, 1991). It is an obligate parasites and induces galls on the leaves, therefore it could be used as a potential method of biological control for S. elaeagnifolium (Cuda et al., 1988; Wapshere, 1988; Sforza, 2007; Kwong and Sagliocco, 2012). The identification of O. phyllobia is based mainly on morphological characters of females and males (Siddiqi, 2000, Subbotin and Riley, 2012). However, due to the large existing number of anguinid nematodes that affect the aerial parts, it is necessary to confirm its identity using more accurate procedures based on their genetic information. In Mexico there is not enough evidence of the presence of this nematode affecting this weed, therefore this research set out to identify by morphotaxonomy, its molecular confirmation and the phylogenetic reconstruction populations of leaf-galling nematode O. phyllobia from Guanajuato, Mexico.
In July, 2015, S. elaeagnifolium plants with deformations or galls on leaves were sampled (Figure 1) in random maize plots (<5 ha) and specific points in urban areas such as highways and roadsides. A total of 12 samples were taken in the municipal area of San Luis de la Paz, Guanajuato, Mexico between coordinates: 21° 16' 24.39" N, 100° 29' 35.17" W, and 21° 18' 33.41" N, 100° 31' 39.32" W. Samples were kept in damp paper to preserve the humidity and the integrity of the specimens and kept in refrigeration at 4 °C ± 2. S. elaeagnifolium leaves with deformities and galls were dissected and incubated in sterile distilled water at 22 °C ± 2 to promote the emergence of the different biological stages (eggs, second-stage juveniles (J2), males and females). For the morphological study, the nematodes (males and females) were transferred onto a drop of water on a microscope slide, were heated with an alcohol burner and mounted in a 2 % water-agar medium (Esser, 1986). The specimens were examined in a Carl Zeiss compound microscope with a phase contrast adapted with a micrometer and a digital camera, measures were taken of the main morphometric characters of males and females. The following characters were considered: L=body length; stylet length, spicule, bursa, and gubernaculum, as well as the De Man ratio: a=body length/maximum body width; b=body length/oesophagous length; b'=body length/distance of the anterior region to the post corpus; c= body length/anus level; c'=tail length/width of body at anus level; V=distance region anterior-vulva; V ' =distance of region anterior to vulva/region anterior to the anus X 100. A morphotaxonomic comparison was carried out of the values reported by Brzeski, 1991; Nickle, 1991; Subbotin and Riley, 2012. To perform the Scanning Electron Microscopy (SEM), males and females were fixed in a formalin-glycerol solution 8:2 for a period of at least 24 h and washed five times with Sorensen's buffer (sodium cacodylate 0.1 M and pH=7.0) every 15 min. In an fume extraction, 500 μl 1 % osmium tetroxide were placed and kept for 2 h; it was poured out and washed another five times with Sorensen's buffer (Eisenback, 1985). Later, they were dehydrated in gradients of ethanol (30, 40, 50, 60, 70, 80, 90, 100 %). The specimens were placed in filter paper envelopes and placed in a Quorum-Emitech K850 critical point dryer (Eisenback, 1986). Once the drying process was completed, the specimens were placed on discshaped sample carriers and observed under a Zeiss Sigma VP scanning electron microscope.

Figure 1 Symptoms caused by the leaf-galling nematode Orrina phyllobia in silver-leaf nightshade Solanum elaeagnifolium. (A), (B) and (C) leaf distorsion and galling; (D) Galling in inflorescence.
The DNA extraction from individual J2, male and female specimens was carried out using the method proposed by Williams et al., 1992 and Thomas et al., 1997 using the specimens previously analyzed by morphotaxonomy. The nematodes were placed individually in 10 μl of lysis buffer solution (10 mM Tris-HCl, pH 8.0, 2.5 mM MgCl2, 50 mM KCl, 0.45 % Tween 20; 0.05 % gelatin) over a coverslip. The specimen was macerated with the tip of a micropipette with the aid of a stereoscopic microscope and the buffer extraction-nematode solution was mixed. The solution was recovered and transferred into a 200 μl microcentrifuge tube and frozen at -40 °C for 30 min. After the time of freezing, samples were incubated at 65 °C for 1 h, shaking at least once, and then adding 0.1 μl of Proteinase K (60 μg/m) in the final 10-15 min. It was incubated at 95 °C for 15 min, to make the Proteinase K inactive, and stored at -20 °C. For the polymerase chain reaction (PCR), two molecular markers were amplified: 1. The region of the ITS1-5.8S-ITS2 of the rDNA using the primers AB28 (5'-GTTTCCGTAGGTGAACCTGC-3') (Joyce et al., 1994) and TW81 (3'-ATATGCTTAAGTTCAGCGGGT-5') (Howlett et al., 1992). 2. The amplification of the expansion segments D2-D3 of gene 28S with the primers D2A (5'-ACAAGTACCGTGAGGGAAAGTTG-3') and D3B (3'-TCGGAAGGAACCAGCTACTA-5') (Nunn, 1992). The PCR reaction for primers AB28 and TW81 was carried out in a final volume of 50 μl with the following components: 5 μl of PCR buffer solution PCR 10X, 3 μl of MgCl2 50 mM, 1 μl mix of DNTP's 10 mM, 2 μl of each primer 10 μM, 0.5 μl of Taq polymerase 5 U/μl, 5 μl of DNA extraction, and 31.5 μl of H2O, free of DNAses and RNAses. The thermocycling conditions included an initial stage of denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 0.45 min, 57 °C for 0.30 min, and 72 °C for 1.30 min, finishing with one cycle at 72°C for 5 min (Skantar et al., 2007). The PCR reaction for primers D2A and D3B included 5 μl of PCR buffer solution 10X, 3 μl of MgCl2 50mM, 1 μl mix of DNTP's 10mM, 2 μl of each initiator 10 μM, 0.5 of μl Taq polymerase 5U/μl, 5 μl of DNA extraction ADN and 31.5 of H2O, free of DNAses and RNAses. The conditions for the amplification were as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 95 °C for 0.45 min, 55 °C for 0.45 min, and 72 °C for 1 min, and finally, one cycle at 72 °C for 5 min (Vovlas et al., 2011). An i-Cycler (BIORAD) thermocycler was used for all reactions. The amplified products were analyzed by horizontal electrophoresis 1.4 % using agarose gels in a buffer solution 1X TAE, dyed with GelRed and visualized in a Gel Doc EZ (BIORAD) photodocumentation. The PCR products were sequenced using Sanger technology, assembled, and edited using the program CodonCode Aligner 4.2.7 (CodonCode Corporation, Centerville, Massachusetts). During the editing of the electropherograms, it was important to manually check for possible errors in the reading of the nitrogenated bases of the DNA sequence, and primers were eliminated.
Once the sequences were obtained for Mexican populations, the search was carried out for homology with sequences of GenBank (NCBI), using the BLAST algorithm to check if the sequences generated of the nematodes that cause galls in S. elaeagnifolium are similar or equal to other sequences reported. The sequences of both molecular markers of the Mexican populations and other GenBank were analyzed by multiple aligment with Clustal W (Thompson et al. 1994). The search for the best substitution model for both markers was performed with Mr Modeltest 2.3 (Nylander, 2004) and PAUP 4.0b10 (Swofford, 1998). The phylogenetic analysis for both markers was carried out using Bayesian inference with MrBayes v3.2 (Ronquist and Huelsenbeck, 2003), with the model GTR+G, with a random starting tree and it was run with the Monte Carlo Markov Chain (MCMC) for 1x106 generations, taking samples at intervals of 100. Two runs were performed for each analysis. After discarding burn-in samples and the evaluation of the convergence, the remaining samples were withheld for later analyses. The topologies were used to generate a 50% majority-rule consensus tree to determine the support values of the clades obtained. For both phylogenies, Radopholus similis (GQ281456) was used for region ITS1-5.8S-ITS2, and Radopholus spp. (HQ823572) for region D2D3 of the gene 28S as out groups. Phylogenetic trees were visualized using Fig Tree (Rambaut, 2009).
The characteristics and morphotaxonomic values of females and males partially agrees with descriptions made by Brzeski (1991), and those reported by Subbotin and Riley (2012) for the leaf-galling nematode Orrina phyllobia (=Ditylenchus phyllobius). Female: Short and straight body 692 (637-785); flat cephalic capsule, in the shape of a capful (Figure 2A and Figure 2D), small stylet 9 (8-9) with large stylet knobs; elongated oesophagus, absence of metacorpus; esophageal gland overlapped with intestine; lateral field with four incisures (Figure 2E); extended ovary, oocytes in one or two rows; short post-uterine sac; pointed tail with rounded tip. Male: Short body and thin 700 (672-737), delicate stylet 9 (8-9) with large knobs; cephalic capsule very similar to that of the female (Figure 2B); short spicule 19.8 (18-21); sub-terminal bursa 58.3 (48-66); tail with projection with pointy ending (Figure 2F). Table 1 shows the morphometric identification of O. phyllobia population in the state of Guanajuato, Mexico and its comparison with the populations of Ditylenchus phyllobius from Arizona, U.S.A., cited by Nickle (1991) and the population from Texas, U.S.A. Brzeski (1991). There are differences in De Man ratios: a (body length/the greatest diameter of the body) and b, (body length/distance of esophageal gland to the anterior end of body when overlapped), since the Mexican population has higher values than those reported. The higher morphometric values can be due to analyzed specimens did not undergo the effect of any chemical fixating agent, since were only anesthetized with heat.
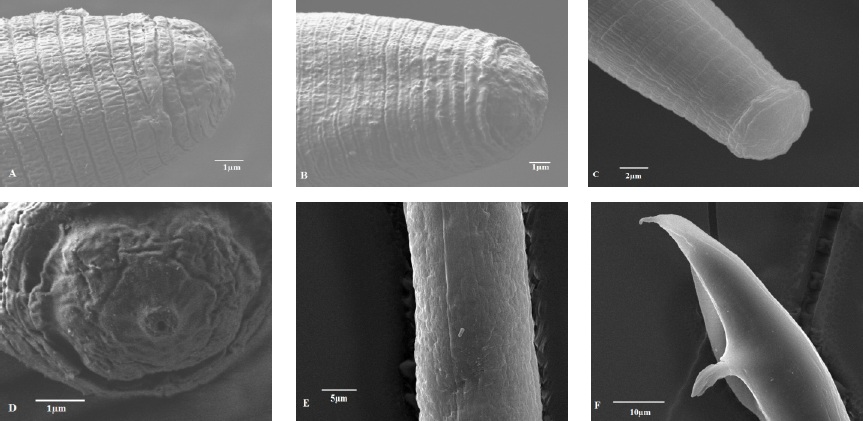
Figure 2 SEM images of Orrina phyllobia; (A) Side view of female's head; (B) Side view of male's head; (C) Side view of head of juvenile J2; (D) Labial region of the female; (E) Lateral fields; (F) Tail of male.
Table 1 Comparative morphometric characteristics of Orrina phyllobia females and males of the population from Guanajuato, Mexico with species reported by Nickle (1991) and Brzeski (1991). All measures are in μm, average (range).
| Carácter |
Orrina phyllobia
(Guanajuato, México) |
Ditylenchus phyllobius
(Nickle, 1991) |
Ditylenchus phyllobius
(Brzeski,1991) |
||||
| Hembra | Macho | Hembra | Macho | Hembra | Macho | ||
| n | 20 | 20 | - | - | - | - | |
| L | 696.2 (637-785) | 700 (672-737) | (590-840) | (670-800) | 684 (592-838) | 750 (672-799) | |
| a | 31.6 (22.5-39) | 32.8 (30-36.8) | (20-32) | (27-34) | 25 (20-32) | 29 (27-34) | |
| b | 7.8 (7.5-10.1) | 8.6 (8.1-9) | (7.4-10.5) | (7.0-9.7) | 8.6 (7.4-10.5) | 8.5 (7-9.7) | |
| b' | 7.7(6.6-9.8) | 8.4 (7.9-8.7) | (4.1 -6.5) | (4.7-7.5) | 5.5 (4.1-6.5) | 5.8 (4.7-7.5) | |
| c | 18.3 (17-19.6) | 20.4 (19.7-22.4) | (11.4-17.6) | (16.3-19.2) | 14.6 (11.4-17.6) | 18.1 (16.3-19.2) | |
| c' | 4.4 (3.5-5.6) | 4 (3.7-4.5) | (2.9-4.5) | (2.5-3.1) | 3.7 (2.9-4.5) | 2.8 (2.5-3.1) | |
| Estilete | 9 (8-9) | 9 (8-9) | (9-10) | (9-10) | (9-11) | (9-10) | |
| Espícula | - | 19.8 (18-21) | - | - | - | 20.5 (19-22) | |
| Bursa | - | 58.3 (48-66) | - | - | - | - | |
| Gubernáculo | - | - | - | - | - | - | |
| V | 79.2 (78-81) | - | (78-85) | - | 81 (78-84) | - | |
| V' | 89.5 (85-92) | - | (85-89) | - | 87 (85-89) | - | |
L=body length; a=body length/maximum body width; b=body length/oesophagus length; b'= body length/distance of the anterior region to the post corpus; c= body length/tail length; c'= tail length/width of body at the level of the anus; V= distance region anterior-vulva; V'= distance of region anterior to vulva/region anterior to anus X 100.
The amplification of ITS1-5.8S-ITS2 of the rDNA and expansion segments D2-D3 of the gene 28S produced a fragment of 850 bp and 900 bp, respectively, based on agarose gel electrophoresis (Figures 3 and 4). The search for homology using BLAST with NBCI sequences for the region ITS1-5.8S-ITS2 of the rDNA shows a 99 % de similarity (E=0.0) with D. phyllobius (AF363112), confirming the identity of the Mexican populations. Regarding the expansion segments D2-D3 of gene 28S, the number of genetic sequences of this group of anguinid nematodes in the NCBI database is very limited, although with the few that exist, a blasting was obtained with 78% similarity with Sequences from Ditylenchus gallaeformans (KF494346), a nematode that causes similar symptoms in weeds in tropical areas of the genus Miconia in Brasil and Costa Rica (Oliveira et al., 2012). Although D. gallaeformans was the species with the highest similarity with sequences from Mexican O. phyllobia populations, the percentage of similarity was very low, and the only relation they have is that they belong to the family of Anguinidae and they cause alterations in the aerial sections of their hosts. The sequences of both molecular markers of the rDNA were deposited in the genetic database of the NCBI (GenBank) with access numbers KT192615 and KT192616 for ITS1-5.8S-ITS2 and KT192617 and KT192618 for the expansion segments D2-D3 of gene 28S as a contribution to the knowledge of Mexican population of this group of nematodes. The Bayesian phylogenetic reconstruction from the region ITS1-5.8S-ITS2 of a multiple alignment that included 37 sequences placed the Mexican O. phyllobia population in an independent clade with a support value of 96.96 % along with the American population of D. phyllobius (=O. phyllobia) AF363112 (Figure 5). This difference is due, in part, to the intraspecific variability between populations of the same species, and on the other hand, the sequenced region of the American O. phyllobia rDNA was partially different since it only included a partial fragment of the gene 18S, the region of the ITS1 and gene 5.8S. Regarding the inference of expansion segments D2-D3 of the gene 28S, multiple alignment included 21 sequences. The data set included 807 nucleotidic characters. Because there are no sequences of this molecular marker in GenBank for this nematode, the Mexican O. phyllobia population and GenBank sequences of other species of anguinids formed independent groups with high support values (Figure 6). It is important to highlight that with both molecular markers, O. phyllobia concentrates on a monophyletic clade (high support value of 100%) and is phylognetically related to Ditylenchus destructor and D. arachidis (ITS1-5.8S-ITS2) and to D. gallaeformans (D2-D3), all members of the Anguinidae family (Figures 5 and 6). These results coincide with those by Douda et al., 2013.
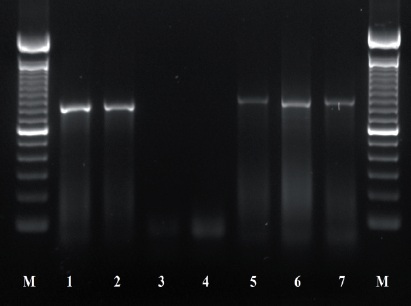
Figure 3 PCR products using primers AB28 and TW81; 1-2: Control (+) Ditylenchus dipsaci; 3-4 control (-) Sterile distilled water; 5-7 population of O. phyllobia. M: Molecular marker 100 bp.
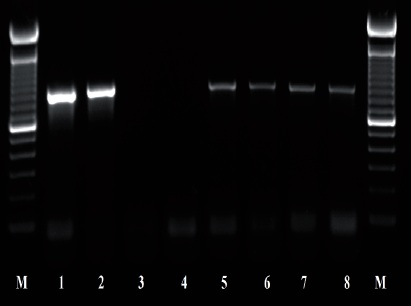
Figure 4 PCR products using primers D2A and D3B; 1-2: Control (+) Ditylenchus dipsaci; 3-4 control (-) Sterile distilled water; 5-8 population of O. phyllobia. M: Molecular marker 100 bp.
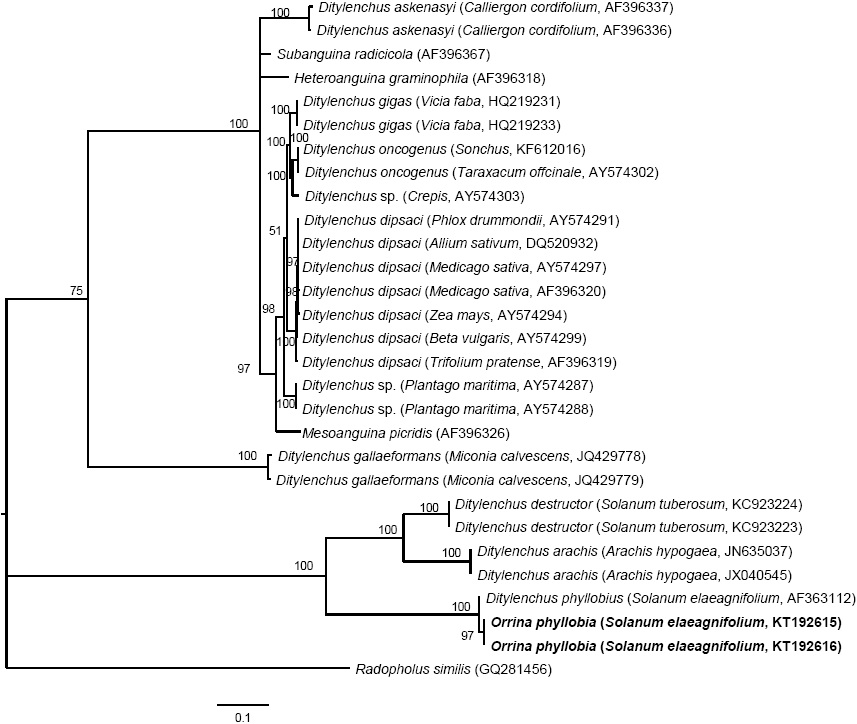
Figure 5 Bayesian inference of nematodes affecting S. elaeagnifolium. A 50% majority-rule consensus tree of the sequences aligned of ITS1-5.8S-ITS2 generated from the complex model: GTR + G with sequences of anguinids deposited in GenBank.
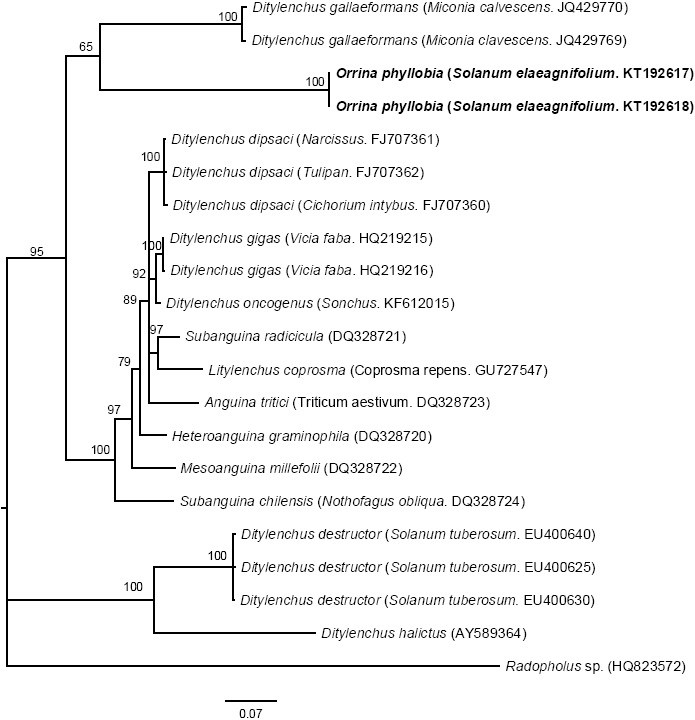
Figure 6 Bayesian inference of sequences of nematodes affecting S. elaeagnifolium. A 50% majority-rule consensus tree of the expansion segments D2-D3 generated from the complex model: GTR + G with sequences of anguinids deposited in the GenBank.
Sequences of the region ITS1-5.8S-ITS2 of the rDNA helped to genetically confirm the identity of Mexican O. phyllobia populations as the causal agent of leaf galling in S. elaeagnifolium, taking as a reference the American D. phyllobius sequence (Powers et al., 2001).
Although there is evidence of O. phyllobia being a prospective biological control agent for the silver-leaf nightshade, there is also the possibility that it affects other species of native solanaceae and other crops such as eggplant Solanum melongena (Field et al., 2009). For future studies, we suggest determining the behavior of O. phyllobia when inoculated in different crop species to determine the host range. The use of taxonomy with an integrated perspective for the identification of nematodes in S. elaeagnifolium was highly important, since it strengthened the benefits of traditional taxonomy. The morphotaxonomic and molecular study helped determine the presence of the foliar nematode Orrina phyllobia attacking silver-leaf nightshade Solanum elaeagnifolium, a weed of economic and ruderal importance in the municipal area of San Luis de la Paz, Guanajuato, Mexico.
Acknowledgements
Our sincere gratitude goes to the Nematology Laboratory of the National Phytosanitary Reference Center (CNRF) of the General Plant Health Department, SENASICA-SAGARPA for having granted the support and helping us carry out and execute this project. To MSc. Jessica Berenice Valencia Luna and MSc. Ariana Guadalupe Robles Zárate of the Area of Electronic Microscopy of the CNRF. This research is part of the Master's Degree thesis of the first author.
REFERENCES
Brzeski M. W. 1991. Review of the genus Ditylenchus Filipjev, 1936 (Nematoda: Anguinidae). Revue de Nematologie 14: 9-59. Disponible en línea: http://horizon.documentation.ird.fr/exl-doc/pleins_textes/fan/40309.pdf [ Links ]
Cuda, J. P., P. E. Parker, R. A. Goodson, and J. L. Gillmore. 1988. Evaluation of Ditylenchus phyllobius (Tylenchida:Anguinidae) as a potential biological control agent for Solanum viarium and Solanum tampicense (Solanaceae). Nematropica 28:107-111. Disponible en línea: http://journals.fcla.edu/nematropica/article/view/64208/61876 [ Links ]
DOF. 1997. Norma oficial mexicana NOM-026-FITO-1995. Por la que se establece el control de plagas del algodonero. México. 13 p. Disponible en línea: http://www.dof.gob.mx/nota_detalle.php?codigo=4893843&fecha=10/09/1997 [ Links ]
Douda O., Marek, M., Zouhar, M. and Rysanek, P. 2013. Insights into the structure and phylogeny of the 28S rRNA expansion segments D2 and D3 of the plant-infecting nematodes from the genus Ditylenchus (Nematoda:Anguinidae). Phytopathologia Mediterranea 52:84-97. Disponible en línea: http://www.fupress.net/index.php/pm/article/viewFile/11334/12198 [ Links ]
Eisenback J. D. 1985. Techniques for preparing nematodes for scanning electron microscopy. pp:79-105. In: Barker, K. R., Carter, G. G. and Sasser, J. N. (eds.). An advanced treatise on Meloidogyne. Volume II: Methodology. A cooperative publication of the Department of Plant Pathology and the United States Agency for International Development. North Carolina State University Graphics, Raleigh, North Carolina, U.S.A. 223p. Disponible en línea: https://www.researchgate.net/publication/234111942 [ Links ]
Eisenback, J. D. 1986. A comparison of techniques useful for preparing nematodes for scanning electron microscopy. Journal of Nematology 18:479-487. Disponible en línea: http://journals.fcla.edu/jon/article/view/69163/66823 [ Links ]
EPPO. 2007. Solanum eleagnifolium Cav. Data sheets on quarantine pests. Bulletin OEPP/EPPO Bulletin 37: 236-245. Disponible en línea: http://www.eppo.int/QUARANTINE/data_sheets/plants/Solanum_elaeagnifolium_DS.pdf [ Links ]
Esser R. P. 1986. A water agar en face technique. Proceedings of the Helminthological Society of Washington 53: 254-255. Disponible en línea: http://bionames.org/bionames-archive/issn/0018-0130/53/254.pdf [ Links ]
Field R. P., Kwong R. M., and Sagliocco, J. L. 2009. Host specificity of Ditylenchus phyllobius, a potential biological control agent of silver-leaf nightshade (Solanum elaeagnifolium Cav.) in Australia. Plant Protection Quarterly 24:141-144. Disponible en línea: http://www.cabdirect.org/abstracts/20093353119.html [ Links ]
Howlett B. J., Brownlee A. G., Guest D. I., Adcock G. J., and McFadden G. I. 1992. The 5S ribosomal RNA gene is linked to large and small subunit ribosomal RNA genes in the oomycetes, Phytopthora vignae, P. cinnamomi, P. megaspera f. sp. glycinae and Saprolegnia ferax. Current Genetics 22:455-461. Disponible en línea: https://www.researchgate.net/publication/21682896 [ Links ]
Joyce S. A., Reid A., Driver F., and Curran J. 1994. Application of polymerase chain reaction (PCR) methods to the identification of entomopathogenic nematodes. Pp:178-187. In: Burnell A. M., Ehlers, R-U, and Masson J-P. (eds). Biotechnology: Genetics of entomopathogenic nematodes-bacterium complexes. Luxembourg: DG XII. European Commission Public. 277p. [ Links ]
Kwong, R. M. and Sagliocco, J.L. 2012. Solanum elaeagnifolum Cav. - silverleaf nightshade. Pp.555-562. In: M. H. Julien, R. E. C. McFadyen and J. M. Cullen. (eds). Biological control of weeds in Australia. CSIRO Publishing. Collingwood, Australia. 619p. [ Links ]
Nickle WR. 1991. Manual of agricultural nematology. CRC Press. 1064p. [ Links ]
Nunn GB. 1992. Nematode molecular evolution. Ph.D. dissertation, University of Nottingham, UK. [ Links ]
Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. Disponible en línea: https://github.com/nylander/MrModeltest2 [ Links ]
Rambaut A. 2009. FigTree. v1.3.1. Available: http://tree.bio.ed.ac.uk/software/. [ Links ]
Robinson A. F., Orr C. C., and Heintz C. E. 1978. Distribution of Nothanguina phyllobia and its potential as a biological control agent of silverleaf nightshade. Journal of Nematology 10: 361-366. Disponible en línea: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2617918/ [ Links ]
Ronquist F. and Huelsenbeck J. P. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. Disponible en línea: http://bioinformatics.oxfordjournals.org/content/19/12/1572.long [ Links ]
Siddiqi M. R. 2001. Tylenchida: parasites of plants and insects. 2nd edition. CABI Bioscience publishing, Wallingford, UK. 835p. [ Links ]
Sforza R. and Jones, WA. 2007. Potential for classical biocontrol of silverleaf nightshade in the Mediterranean Basin. EPPO Bulletin 37:156-162. Disponible en línea: http://dx.doi.org/10.1111/j.1365-2338.2007.01109.x [ Links ]
Skantar A. M., Handoo Z. A., Carta L. K., and Chitwood D. J. 2007. Morphological and molecular identification of Globodera pallida associated with potato in Idaho. Journal of Nematology 39:133-144. Disponible en línea: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2586493/ [ Links ]
Subottin, S. A. and Riley I. T. 2012. Stem and gall forming nematodes. Pp.:521-577. In: Manzanilla-López, R. H. and Marbán-Mendoza, N. (eds.). Practical Plant Nematology. Biblioteca Básica de Agricultura (BBA). Montecillo, Texcoco, Estado de México. México. 883p. [ Links ]
Swofford DL. 1998. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts. Disponible en línea: http://paup.csit.fsu.edu/Cmd_ref_v2.pdf [ Links ]
Thomas W. K., Vida J. T., Frisse L. M., Mundo M., and Baldwin, J. G. 1997. DNA sequences from formalin-fixed nematodes: Integrating molecular and morphological approaches to taxonomy. Journal of Nematology 29:250-254. Disponible en línea: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619797/ [ Links ]
Thompson J. D., Higgins D. G., and Gibson T. J. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673-4680. Disponible en línea: http://www.ncbi.nlm.nih.gov/pubmed/7984417 [ Links ]
Vibrans H. 2009. Malezas de México. CONABIO. Fecha de consulta 08 diciembre del 2015. Disponible en línea: http://www.conabio.gob.mx/malezasdemexico/2inicio/home-malezas-mexico.htm [ Links ]
Vovlas N., Troccoli A., Palomares-Rius J. E., De Luca F., Liebanas G., Landa B. B., Subbotin S. A., and Castillo P. 2011. Ditylenchus gigas n. sp. parasitizing broad bean: A new stem nematode singled out from the Ditylenchus dipsaci species complex using a polyphasic approach with molecular phylogeny. Plant Pathology 60:762- 775. Disponible en línea: http://www.readcube.com/articles/10.1111%2Fj.1365-3059.2011.02430.x [ Links ]
Wapshere, A. J. 1988. Prospects for biological control of silverleaf nightshade, Solanum elaeagnifolium, in Australia. Australian Journal of Agricultural Research 39:187-97. Disponible en línea: doi:10.1071/AR9880187 [ Links ]
Williams B. D., Schrank B., Huynh C., Shownkeen R., and Waterston R. H. 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequencetagged sites. Genetics 131:609-624. Disponible en línea: http://www.genetics.org/content/genetics/131/3/609.full.pdf [ Links ]
Received: January 11, 2016; Accepted: March 28, 2016











 text in
text in 


