http://zoobank.org/urn:lsid:zoobank.org:pub:E74CB8AA-FF75-49FD-97D1-B8B0519C7E5D
Introduction
Hymenolepis Weinland, 1858 includes species that feature an unarmed scolex with a rudimentary rostellar apparatus, anastomoses connecting ventral excretory canals, saccate uterus and testes arranged triangularly or in a transversal line. There are 10 species known to occur in North America which infect sciurid, arvicoline, geomyid and neotomine rodents (Gardner et al., 2020; Hoberg et al., 2016; Makarikov et al., 2015). In addition, the cosmopolitan H. diminuta Weinland, 1858 has been recorded throughout the continent infecting humans and both synanthropic and wild rodents (Coggins & McDaniel, 1975; Faulkner & Lochmiller, 2000; Harkema, 1946; Harkema & Kartman, 1948; Mollhagan, 1978; Seidenberg et al., 1974).
Although the majority of Hymenolepis species are known to infect only their definitive host, two species occur in unrelated sympatric mammals. These include H. citelli (McLeod, 1933) and H. folkertsiMakarikov, Nims, Galbreath and Hoberg, 2015; the latter was documented from a variety of unrelated species of cricetid, geomyid, heteromyid and sciurid rodents distributed across the United States (Gardner, 1985; Gardner et al., 2020; Hoberg et al., 2016; Makarikov et al., 2015; Pfaffenberger et al., 1985; Riley & Shannon, 1922; Smith et al., 1953). Recently, the use of mitochondrial genes has assisted in establishing the species identity of individual tapeworms identified as H. folkertsi collected across the vast geography of North America and from a wide diversity of mammals. By analyzing sequence divergence, Hoberg et al. (2016), established inter-population genetic structure and related this to the characterization of intraspecific morphological variability of diagnostic characters. They also determined that the genetic distance among individuals of H. folkertsi across the distribution of the species oscillates between 1.9 and 2.7% (Hoberg et al., 2016). A similar approach has been used to evaluate the intraspecific genetic diversity in species of the hymenolepidid Arostrilepis Mas-Coma and Tenora, 1997 (Makarikov et al., 2020) and provide identifications for human-dwelling hymenolepidids (Nkouawa et al., 2016).
Ongoing biodiversity monitoring of small mammals and their associated ecto- and endo-parasite faunas on the Konza Prairie Biological Station (KPBS), and Long-Term Ecological Research (LTER) site, located in northeastern Kansas, USA, reflects 6 consecutive years of both mark-recapture data and comprehensive mammal and parasite voucher collection (Galbreath et al., 2019; Hope, 2019). These integrated surveys are performed with the purpose of documenting the distributional ecology, population biology and co-evolutionary history of these communities, as they occur across a landscape scale experimental manipulation of land-use treatments. These treatments include a factorial design of controlled wildfire and grazing regimes that have been continuously managed for > 40 years of ecological experimentation focused on understanding drivers of grassland resilience. The KPBS is located in the Flint Hills ecoregion and supports the largest remaining tract of native tallgrass prairie through the central Great Plains. Experimental manipulation, especially through wildfire suppression has resulted in a mosaic landscape of native prairie, shrubland communities, and late-seral woodlands. Small mammals were sampled in multiple habitats reflected by variable fire return intervals. Although most of this site is located on terraced limestone and has never been suitable for agriculture, the regional impacts of changing land use has profoundly altered most tallgrass prairie habitats (Briggs et al., 2005), associated mammalian community structure (Bruckerhoff et al., 2021), and ecosystem function (Ratajczak et al., 2016). To date, the lack of a detailed species level inventory of parasites in this locality precludes any knowledge of their biological role in this ecosystem.
Here we report some findings from our efforts to identify the species of tapeworms present in tallgrass prairie rodents and propose the description of a new species recovered from hispid cotton rats Sigmodon hispidus Say and Ord, 1825, eastern woodrats Neotoma floridana (Ord, 1818), and prairie voles Microtus ochrogaster (Wagner, 1842). Our description is based on the examination of the morphological features that characterize these parasites as well as their molecular identity based on mitochondrial genes.
Materials and methods
Mammals were trapped using Sherman® live traps (H.B. Sherman, Tallahassee, FL). Subsequently they were euthanized under approved protocols (Hope IACUC #3579, #4055) and processed to preserve all specimen parts (Galbreath et al., 2019) for permanent archive in the Museum of Southwestern Biology, University of New Mexico (http://arctos.database.museum/). The trapping design used at the Konza Prairie Biological Station (KPBS), and Long-Term Ecological Research (LTER) site, (Kansas, USA, 39.07264° N, 96.57818° W, 416 m elevation) is detailed in the data report Universally available (Hope et al., 2019). For parasite preservation, the digestive system was cut open and the tapeworms present were removed. Worms were washed and killed with hot water to induce their relaxation. Specimens were preserved in 70% ethanol, washed, postfixed in 4% formaldehyde and stained in Semichon’s paracarmine to be mounted permanently in Canada Balsam. Specimens identified as H. diminuta collected from hispid cotton rats from Oklahoma were borrowed from the National Parasite Collection (USNM1382519 = USNPC87319) and used for comparison. All measurements are in micrometers (µm), unless otherwise noted. Type material was deposited at Museum of Southwestern Biology (MSB), Division of Parasites, University of New Mexico, Albuquerque, New Mexico, USA; Harold W. Manter Laboratory of Parasitology (HWML), University of Nebraska, Lincoln, Nebraska, USA, and Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City, Mexico.
Prior to the staining and mounting of specimens, fragments of a few contiguous mature proglottids were excised and used for the extraction of genomic DNA using a commercial kit (Qiagen, Valencia CA), whereas the rest of the specimen was processed as described above for morphological examination. The mitochondrial NADH dehydrogenase subunit 1 (Nad1) gene was amplified using primers and thermal profiles developed and described elsewhere (Haukisalmi et al., 2010; Morgan & Blair, 1998). The resulting amplicons were cleaned using ExoSAP (GE Healthcare, Cleveland, OH) and submitted to a commercial facility to be sequenced using Sanger chemistry (Eurofins Inc., Lexington, Kentucky). Sequences were assembled in Sequencher 3.5 (Gene Codes Corp. Ann Arbor, Michigan), and aligned and trimmed with available homologous sequences from hymenolepidids and its putative related groups Arostrilepis, Staphylocystis Villot, 1877 and Staphylocystoides Yamaguti, 1959 using Clustal W (Sievers et al., 2011) with default settings. Accession numbers of the sequences used are related in Table 1. Resulting alignments had a size of 712 bp. Following alignment, the model of evolution GTR was selected using the best fit model according to the corrected Akaike Information Criterion as implemented in JModeltest (Posada, 2008). The posterior probability of branches was calculated using Bayesian inference as implemented in Mr. Bayes v3.2.7 using the CIPRES portal (Miller et al., 2010). Trees were visualized using FigTree v1.4.3 (Rambaut, 2012). Genetic distances were estimated with PAUP*.
Table 1 List of specimens and sequences used to analyze the phylogenetic position of Hymenolepis ackerti.
| Species | ND1 GenBank No. | Collection number | Locality | Host species |
|---|---|---|---|---|
| Arostrilepis beringiensis | KM516216 | Irkutsk, Russia | Lemmus sibiricus | |
| Hymenolepididae | JQ950694 | MHNGE-INVE 79505 | Hualaihue, Chile | Sephanoides sephaniodes |
| Hymenolepis ackerti | OP649625 | MSB: Para: 35315 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649626 | HWML118076 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649633 | MSB: Para: 35316 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649634 | HWML118077 | Kansas, USA | Neotoma floridana |
| Hymenolepis ackerti | OP649632 | MSB: Para: 35317 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649631 | MSB: Para: 35318 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649630 | MSB: Para: 35319 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649629 | MSB: Para: 35314 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649628 | MSB: Para: 35313 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis ackerti | OP649627 | HWML118078 | Kansas, USA | Sigmodon hispidus |
| Hymenolepis diminuta | AF314223 | N/A | Lab, Michigan | Lab isolate |
| Hymenolepis diminuta | AP017664 | N/A | Lab, Japan | Lab isolate |
| Hymenolepis diminuta | LR536429 | N/A | Lab strain WMS-il1, Poland | Lab Isolate |
| Hymenolepis diminuta | HM149290 | N/A | Madagascar | Rattus rattus |
| Hymenolepis diminuta | HM149291 | N/A | Canary Islands, Spain | Rattus rattus |
| Hymenolepis folkertsi | OP649624 | 35314 | Kansas, USA | Peromyscus leucopus |
| Hymenolepis folkertsi | KX782315 | N/A | Pennsylvania, USA | Peromyscus leucopus |
| Hymenolepis folkertsi | KX792199 | MSB: Para: 23512 | Idaho, USA | Tamias amoenus |
| Hymenolepis folkertsi | KX792195 | MSB: Para: 23503 | Michigan, USA | Peromyscus leucopus |
| Hymenolepis nana | AP017666 | N/A | Lab, Japan | Lab isolate |
| Hymenolepis sp. C31 | HM149299 | N/A | Madagascar | Eliurus tanala |
| Hymenolepis sp. | HM149298 | N/A | Madagascar | Tenrec ecaudatus |
| Hymenolepis sp. U45 | HM149292 | N/A | Kazakhastan | Microtus socialis |
| Hymenolepis sp. AY0 | HM149295 | N/A | Wyoming, USA | Thomomys sp. |
| Hymenolepis sp. BT0 | HM149296 | N/A | Victoria, Australia | Rattus fuscipes |
| Hymenolepis sp. CA9 | HQ589353 | N/A | South Korea | Apodemus agrarius |
| Hymenolepis sp. U9 | HM149297 | N/A | Turkey | Apodemus sylvaticus |
| Hymenolepis sp. Z39 | HM149289 | N/A | Bosnia and Herzegovina | Microtus subterraneus |
Description
Genus Hymenolepis Weinland, 1858
Hymenolepis ackerti n. sp. (Figs. 1, 2)
http://zoobank.org/urn:lsid:zoobank.org:act:170E75EE-96A9-40B9-B814-3A54CF26D6BE
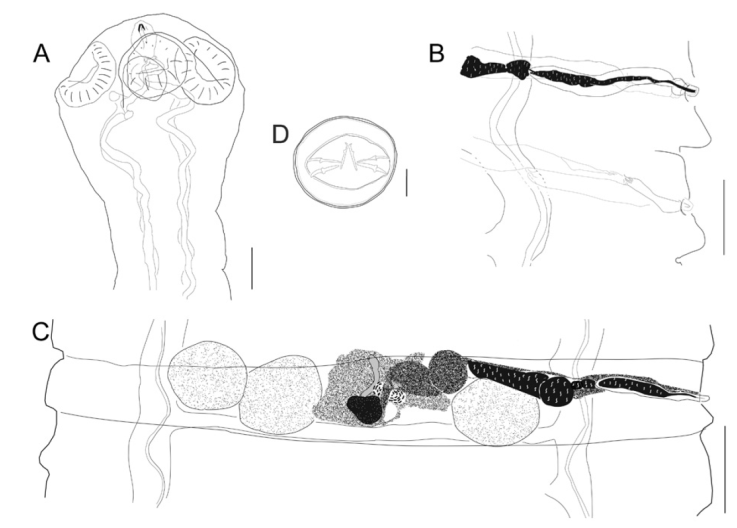
Figure 1 Hymenolepis ackerti n. sp. A, Scolex showing anterior portion of excretory canals that do not anastomose or reach the anterior canal. Scale bar = 100 μm; B, detail of genitalia showing genital atrium, cirrus sac containing both cirrus and internal seminal vesicle, and distal end of the external seminal vesicle. The connection of the vagina with the seminal receptacle is shown in the lower proglottid. Scale bar = 100 μm; C, dorsal view of a mature proglottid showing the relative position of the ovary, seminal receptacle, location of testes, sigmoidal external seminal receptacle, and cirrus sac. Scale bar = 200 μm; D, uterine egg featuring hooks in onchosphere. Scale bar = 25 μm.
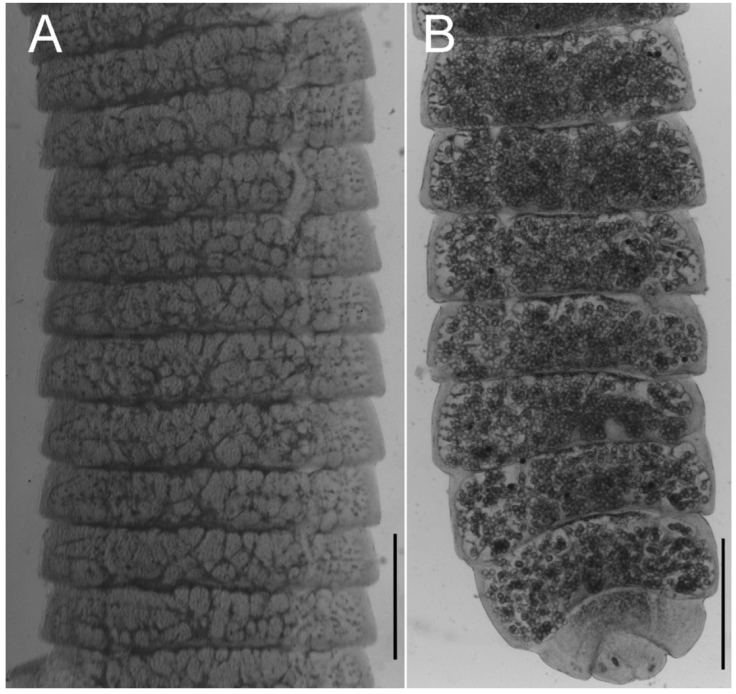
Figure 2 Micrographs of uterine development of Hymenolepis ackerti n. sp.(holotype). A, Ventral view of proglottids featuring developed uterine branches in the space between excretory ducts, scale bar = 500 µm; B, ventral view of terminal gravid proglottids, showing all uterine branches filled with eggs, scale bar = 1 mm.
Based on 10 specimens, measurements of the holotype in parenthesis. Scolex unarmed, semispherical, 417-570 (548) long by 438-567 (472) wide (Fig. 1A). Rostellar capsule 100-138 (100) long by 66-87 (77) wide, thin walls not reaching the posterior margin of suckers; anterior canal or apical organ 31-36 (31). Osmoregulatory canals terminate in scolex, near posterior margin of rostellar capsule (Fig. 1A). Suckers unarmed (n = 40), slightly oval, 112-192 (126 - 169) long by 103-182 (145-160) wide, endowed with thick muscular walls. Neck 293-423 (423) long, conspicuously narrower than scolex (Fig. 1A). Relatively small fully developed strobila, length 40.4-106.6 mm (106.6, n = 8), maximum strobilar width 1,084-2,207, (2,207 mm) at level of newly gravid proglottids. Ventral excretory canals 62-93 (93) wide, connected with transversal anastomoses. Dorsal excretory canal 7-10 (9) wide. Development of proglottids protandrous; proglottids feature non-alternating dextral genital pores (Fig. 1B).
Proglottids anapolytic wider than long; strobilar margins craspedote, with conspicuous intersegmental divisions between mature and gravid proglottids. Mature proglottids 97-225 (175-225, n = 64) long by 1,260-1,771 (1,587-1,771, n = 64) wide. Genital atrium simple, infundibular, located at anterior end of proglottid margin 18-29 (18). Cirrus sac pyriform and muscular, relatively long 142-266 (142-256, n = 128) by 32-61 (32-61, n = 82); antiporal portion of cirrus sac occasionally reaches ventral excretory canal, yet never crosses longitudinal osmoregulatory canals (Fig. 1B, C). Cirrus claviform, armed with minute spines 73-101 (78-97, n = 8). Internal seminal vesicle oval, 73-186 (73-166, n = 42) more than half of cirrus sac length (Fig. 1B, C); external seminal vesicle fusiform 166-235 (205-273, n = 36), dorsoventrally undulating and distinctly shorter than seminal receptacle (Fig. 1B, C). Genital ducts cross dorsally both ventral and dorsal osmoregulatory canals. Usually 3 testes, 1 poral, 2 antiporal; 4 or 5 testes rarely occur. Testes oval, somewhat linearly arranged 88-205 (147-205, n = 120) long by 123-273 (161-245, n = 120) wide; poral testis separated from antiporals by female genitalia (Fig. 1C). Vaginal opening ventral to cirrus opening, copulatory part of vagina 75-107 (75-88, n = 16) long (Fig. 1B). Fusiform seminal receptacle, 520-714 (608-710, n = 24) (Fig. 1C). Ovary median, fan-shaped, 45-159 (85-106, n = 46) long by 108-304 (169-188, n = 41) wide, occasionally overlapping proximal poral testis. Vitellarium lobed, postovarian 50-169 (80-169, n = 40) long by 72-200 (72-143, n = 40) wide.
Uterus develops laterally from the central part of the proglottid and elongates to fill the space between the excretory canals and the space closer to lateral margins (Fig. 2A, B); saccular projections dorsally directed. In gravid proglottids the saccular uterus fills the entirety of the proglottid, eggs occupy first the space between excretory canals and subsequently eggs fill the marginal spaces in terminal proglottids (Fig. 2B). Gravid proglottids 275-803 (548-803, n = 76) long by 1,084 - 2,207 (1,851-2,207, n = 76) wide.
Outer coat of eggs usually spherical 38-55 (35-36, n = 50) long by 38-56 (40-45, n = 50) wide, surface endowed with uniform embossment; onchosphere 18-33 (20-33) long by 19-37 (22-37) wide. Embryophore 25-36 (25-29) long by 34-40 (34-35) wide (Fig. 1D). Pair of slender median hooks, 8-13 (9-12, n = 15); anterolateral hooks relatively robust 11-16 (11-15, n = 43). Terminal or last proglottids contain no eggs (Fig. 2D).
Taxonomic summary
Symbiotype: hispid cotton rat, Sigmodon hispidus (Say and Ord).
MSB: Mamm: 330681.
Site of Infection: small intestine.
Type locality: USA, Kansas, Konza Prairie Biological Station (KPBS), and Long-Term Ecological Research (LTER) site, 39°4’22” N, 96°34’41” W, 416 m elevation.
Specimens deposited: holotype (MSB: Para: 35308). Paratypes MSB: Para: 35309, HWML216883, HWML118076, CNHE11734. Vouchers: MSB: Para: 35306-35307, 35310-35311; 35313-35319; HWML118077 and 118078, CNHE11735.
Other hosts: prairie vole, Microtus ochrogaster (Wagner) and the eastern woodrat, Neotoma floridana (Ord)
Specimens examined: Neotoma floridana MSB: Mamm: 330678, Microtus ochrogaster MSB: Mamm: 331573 and Sigmodon hispidus MSB: Mamm: 305504, MSB: Mamm: 305616, MSB: Mamm: 330605, MSB: Mamm: 330663, MSB: Mamm: 331560, MSB: Mamm: 331542, MSB: Mamm: 331563, MSB: Mamm: 331584, MSB: Mamm: 331590, MSB: Mamm: 332155.
NCBI GenBank numbers: OP649625 - OP649634, OQ407494 - OQ407496
Etymology: the species is named after J. E. Ackert, former president of the American Society of Parasitologists and head of the former Zoology Department at Kansas State University.
Remarks
Hymenolepis ackerti is unique among all described species of this genus present in North America because of the extremely large and semispherical scolex, the small embryo hooks and the combination of a relatively large cirrus, relatively large seminal receptacle, small eggs and the disposition of the rostellar capsule that barely reaches the anterior margin of the suckers. The rostellar capsule of H. ackerti is completely embedded in the scolex, which allows the differentiation of this species from H. folkertsi, H. pitymi Yarinsky, 1952, and H. tualatinensisGardner, 1985; further, all 3 of these species feature testes arranged in a triangle, as opposed to the linear arrangement characteristic of H. ackerti.
In some individuals the seminal receptacle of H. ackerti, is relatively elongated and reaches the center of the proglottid. This is notable because this proportion is seen in only four other species present in the New World, which include H. cratogeomys, H. diminuta, H. scalopi, and H. weldensis Gardner and Schmidt, 1988. However, the eggs and hooks present in the embryo of H. ackerti are typically smaller than the homologous structures in those four species. In particular, H. ackerti shares six characteristics with H. weldensis, namely the configuration of the rostellar capsule, the testicular arrangement, the size of ovaries, length of seminal receptacle, size of vitelline glands and size and extension of cirrus sac. However, both species can be differentiated by the size of the eggs, onchosphere and embryonic hooks, being larger in H. weldensis.
Compared to H. robertrauschi, individuals of H. ackerti feature relatively smaller eggs and embryos, yet rostellar capsule and seminal receptacle are at least twice as long in H. ackerti. The larger size of the scolex and the smaller hooks in the onchopsphere can be used to differentiate H. ackerti from the rest of the North American species. The scolex of H. ackerti is almost twice as wide as the homologous structure of H. citelli, H. cratogeomyos, H. gomydis and H. scalopi (Table 2). Finally, only the eggs of H. pitymi appear to be smaller than those in H. ackerti, yet both species can be differentiated based on the arrangement of the testes, linear in the latter and triangular in the former. The relative size of the hooks in the embryo are similar to those present in H. diminuta; however, the testes and the eggs of H. ackerti are slightly larger, whereas the cirrus sac is usually longer in H. diminuta.
Table 2 Comparative measurements of relevant characters used in the taxonomy of Hymenolepis species from North America.
| H. ackerti n. sp. | H. diminuta (Rudolphi, 1819) | H. citelli (McLeod, 1933) | H. crateogeomyosGardner, Dursahinan, Campbell and Rácz, 2020 | H. tualatinensis Gardner, 1985 | H.weldensis Gardner and Schmidt, 1988 | H. geomydis Gardner and Schmidt, 1988 | H. folkertsi Makarikov, Nims, Galbreath and Hoberg 2015 | H. scalopi Schultz, 1939 | H. pitymi Yarinsky 1952 | H. robertrauschi Gardner, Luedders and Duszinski, 2014 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Scolex with rostrum-like projection | No | No | No | No | Yes | No | No | Yes | No | Yes | No |
| Strobila length (mm) | 106.57 | 183 | 150 | 88-129 | 24-210 | 111-165 | 72-168 | 99-116 | 100-200 | 19.6 | 42.4 - 83.4 |
| (40-51) | |||||||||||
| Strobila width (mm) | 1.52 | 2.38-2.56 | 2.8 | 2.72-3.41 | 1.75 | 0.82-0.94 | 1.98-3.3 | 1.71-1.85 | 2.57 | 750 | 1.18 - 2.54 |
| (1.69-1.94) | |||||||||||
| Scolex width | 472 | 286-296 | 245 | 209-227 | 92-167 | 126-288 | 194-245 | 168 | 170 - 232 | 199 - 257 | |
| (438-568) | |||||||||||
| Sucker size | 126 - 169 × 145 - 160 | 96-108 | 113 × 87 | 84-89 × 109-117- | - | - | 92-124 × 65-94 | 93-102 × 70-86 | 35-62 × 86-92 | 119 - 164 × 82-95 | |
| (111-184 × 121-182) | |||||||||||
| Rostellar pouch size (length × width) | 100 × 77 (115-138X66-87) | 94-108 × 44-52 | 55-59 | 51-61 long | - | - | 67 × 43 long | 121 | 50 long | ||
| Mature proglottid size | - | - | - | - | - | - | - | 90-150 × 810-1030 | |||
| Testes size (length × width) | 93-200 × 131-272 | 118-220 × 40-48 | 143 × 113 | 141-190 × 95-121 | 63-141 × 54-141 | 92-166 | 55-180 × 81-180 | 75-112 × 44-62 | 101-123 × 87-113 | ||
| Testicular arrangement | Linear | Linear | Linear | Linear | Triangular | Linear | Triangular | Triangular | Linear | Triangular | Triangular |
| Cirrus-sac size | 143-256 × | 240-300 × | 157 long | 121-145 × | 56-150 × | 149-194 × | 80-160 × | 138-154 × | 125-159 × | 79 long | 147 - 233 × |
| (length × width) | 32-61 | 28-48 | 45-57 | 26-49 | 34-51 | 36-67 | 30-39 | 42-49 | 33-61 | ||
| Cirrus size | 78-97 | 38-47* | - | __- | - | - | - | 45-58 × 7-10 | 84 - 157 | ||
| (72-102) | |||||||||||
| Cirrus armature | minute spines | minute spines | - | armed | minute spines | minute spines | minute spines | minute spines | minute spines | ||
| Ovary width | 169-188 | - | 168-203 | 96-216 | 90-293 | 180-494 | 147-217 | 193-220 | 97-116 | 130 - 297 | |
| (108-246) | |||||||||||
| Vitellarium size (length × width) | 80-169(50-89 × 87-167) | - | - | 45-53X62-80 | 34-109 × 37-132 | 50-112 × 54-106 | 61-137 × 101-209 | 30-57 × 55-98 | 68-83 × 78-92 | 9,46-118× 45 - 102 | |
| Seminal receptacle size (length × width) | 608 - 710 | 71-540×28 - 200 | 220 - 229 × 37-46 | 746-919 × 75-93 | 48-169 × 23-70 | 175-552 × 43-148 | 99-369 × 59-108 | 275-365 × 37-62 | 533-597 × 60-68 | 183 - 210 × 42-51 | 190 - 246 |
| Egg size (length × width) | 32-36 × 40-45(38-55X38-56) | 56-60 × 63-66 | 59-65 × 78-86 | 62-94 × 40-63 | 57-89 × 42-68 | 70-81 × 67-77 | 76-85 × 72-83 | 46-56 × 57-69 | 65-80 × 49-62 | 28×31 | 57-76×44 - 58 |
| Oncosphere size (length × width) | 20-33 × 22-37(18-32 × 19-33) | 23-25 × 28-31 | - | 23-49 | 38-45 × 38-40 | 38-50 × 34-43 | 22-26 × 26-35 | 23×20 | 40-57×31 - 53 | ||
| Embryonic Hook size | 11-16 | 14-15 | 16-20 | 14-20 | 17-20 | 13-16 | 16-20 | 17-18 | 16-21 | 12-18 | 15.7 - 20.5 |
Genetic screening of individuals collected from different species of mammals
The monophyly of H. ackerti is supported by their phylogenetic placement according to the analyses of ND1 mtDNA locus herein analyzed (Fig. 3). In the resulting topologies, H. ackerti is not the sister species for either H. diminuta nor for H. folkertsi, the other species present in North America for which comparative GenBank sequences were available. All sequenced specimens of H. ackerti differ by one or two single point mutation representing three haplotypes of ND1, their intraspecific genetic distance oscillates between 0.15 and 0.3% (Table 3).
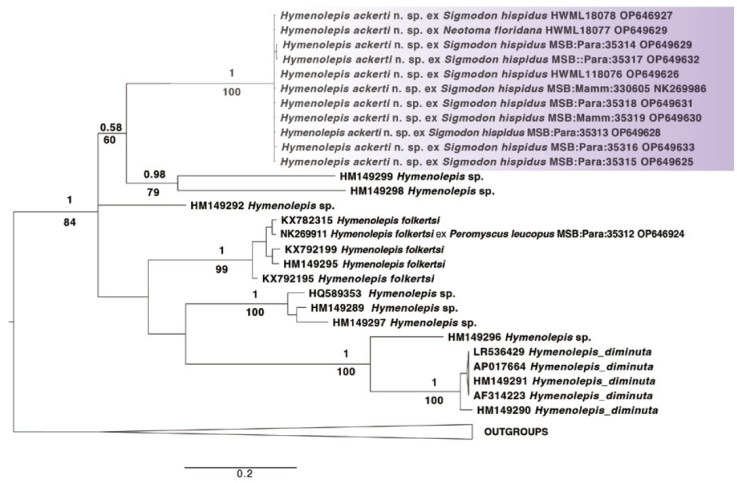
Figure 3 Phylogenetic reconstruction showing the position of Hymenolepis ackerti relative to available sequences of the mtDNA ND1 locus for hymenolepidids. The values atop each node represent posterior probabilities (Bayesian inference) and values below represent Bootstrap support based on 1,000 replicates.
Table 3 Genetic distance for specimens of Hymenolepis folkertsi and Hymenolepis ackerti based on the mt locus ND1. Fractions below the diagonal represent the genetic distance based on the Jukes Cantor model. The numbers above the diagonal represent the absolute number of point mutations among the compared sequences.
| Taxon, GenBank, host, collection number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | H. folkertsi KX782315 ex Peromyscus leucopus | - | 12 | 12 | 15 | 4 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 |
| 2 | H. folkertsi KX792199 ex Tamias amoenus MSB: Para: 23512 | 0.019 | - | 8 | 17 | 12 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 3 | H. folkertsi HM149295 ex Thomomys sp. | 0.019 | 0.012 | - | 17 | 12 | 104 | 104 | 104 | 104 | 104 | 104 | 104 | 104 | 104 | 104 | 104 |
| 4 | H. folkertsi KX792195 ex P. leucopus MSB: Para: 23503 | 0.024 | 0.027 | 0.027 | - | 15 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 |
| 5 | H. folkertsi OP649624 ex. P. leucopus MSB: PARA: 35312 | 0.006 | 0.019 | 0.019 | 0.024 | - | 97 | 97 | 97 | 97 | 97 | 97 | 97 | 97 | 97 | 97 | 97 |
| 6 | H. ackerti OP649627 ex S. hispidus HWML118078 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | - | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 7 | H. ackerti OP649634 ex N. floridana HWML118077 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | - | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 8 | H. ackerti OP649626 ex S. hispidus HWML118076 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | - | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 9 | H. ackerti OP649629 ex S. hispidus MSB: PARA: 35314 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0.002 | 0.002 | 0.002 | - | 1 | 1 | 0 | 1 | 1 | 2 | 1 |
| 10 | H. ackerti OP649630 ex S. hispidus MSB: PARA: 35319 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | 0 | 0.002 | - | 0 | 1 | 0 | 0 | 1 | 0 |
| 11 | H. ackerti NK269986 ex S. hispidus MSB: Mamm: 330605 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | 0 | 0.002 | 0 | - | 1 | 0 | 0 | 1 | 0 |
| 12 | H. ackerti OP649632 ex S. hispidus MSB: PARA: 35317 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0.002 | 0.002 | 0.002 | 0 | 0.002 | 0.002 | - | 1 | 1 | 2 | 1 |
| 13 | H. ackerti OP649631 ex S. hispidus MSB: PARA: 35318 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | 0 | 0.002 | 0 | 0 | 0.0016 | - | 0 | 1 | 0 |
| 14 | H. ackerti OP649628 ex S. hispidus MSB: PARA: 35313 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | 0 | 0.002 | 0 | 0 | 0.0016 | 0 | - | 1 | 0 |
| 15 | H. ackerti OP649633 ex S. hispidus MSB: PARA: 35316 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0.002 | 0.002 | 0.002 | 0.003 | 0.002 | 0.002 | 0.0031 | 0.002 | 0.002 | - | 1 |
| 16 | H. ackerti OP649625 ex S. hispidus MSB: PARA: 35315 | 0.161 | 0.173 | 0.181 | 0.161 | 0.167 | 0 | 0 | 0 | 0.002 | 0 | 0 | 0.0016 | 0 | 0 | 0.002 | - |
Discussion
Cestode specimens collected from hispid cotton rats in the last 75 years were usually identified as H. diminuta (Coggins & McDaniel, 1975; Faulkner & Lochmiller, 2000; Harkema, 1946; Harkema & Kartman, 1948; Mollhagan, 1978; Seidenberg et al., 1974). From these, only few specimens collected from Oklahoma were deposited in scientific collections as verifiable voucher specimens. Examination of these specimens, USNM1392519 = USNPC87319, revealed they cannot be diagnosed as H. diminuta, based on the fact that they feature an everted armed rostellum, not typical for the species.
Relative to the intraspecific morphological variability, it must be noted that specimens of H. ackerti feature relatively short proglottids and most organs are arranged in the same dorsoventral plane. A similar situation occurs with the position of the vitellaria relative to the ovary. Further, the length of the strobila of some specimens infecting hispid cotton rats is similar to the strobilar size of H. diminuta (about 180 mm). However, this is quite variable and it appears that it depends on the number of individuals infecting a single host. In hosts infected by only 2 parasites, worms measuring up to 107 mm (MSB: Mamm: 330681), yet for high intensity infections (~ 30 individuals) worms seldom exceeded 51 mm (MSB: Mamm: 305616).
Relative to the linear testicular arrangement of H. ackerti, it must be noted that in most of the specimens the internal testis is slightly more posterior in position compared with both poral and antiporal testes. This is a very similar arrangement as the one seen in H. weldensis and H. robertrauschi. Interestingly, the testicular arrangement in the latter is described as triangular, whereas the arrangement in the former is described as linear. Perhaps the only three species featuring an authentic triangular arrangement are H. folkertsi, H. pitymi and H. tualatinensis, which coincidentally also feature a rostral projection of the rostellar capsule. Yet, even in this group, there is considerable variation in the testicular arrangement, as shown by the clearly triangular arrangement of the type specimens of H. folkertsi (Makarikov et al., 2015), which contrast with the almost linear arrangement seen in conspecific specimens identified here (specimen collected from the prairie vole MSB: Mamm: 331573) and documented subsequently (Hoberg et al., 2016).
The phylogenetic position of parasite specimens hosted by hispid cotton rats from the Konza prairie suggests that these specimens are not conspecific with H. diminuta nor H. folkertsi, other species that occur in North America and are expected to infect a wide variety of rodents. Further, their morphological traits appear to be unique among all species known from North America, suggesting that an endemic species of Hymenolepis infects voles, hispid cotton rats and other rodents through the Great Plains region. Parasites were recovered from rodent hosts sampled from multiple habitat types including open grassland, shrubby habitats, and woodland. However, shrubby and woody habitats yielded the vast majority of specimens, with potential implications for increased intensity of infection through ongoing woody encroachment of the region (Briggs et al., 2005). Recent reevaluation of specimens and new helminthological surveys have resulted in the description of 4 species of hymenolepidids in the continental United States in recent years (Gardner et al., 2014; Makarikov et al., 2020, 2015). We posit that increasing sampling effort of tapeworms from rodents may yield a modest increase in the number of species described. Nevertheless, the completion of this effort will ensure a better functional understanding of the relationship between parasites, their arthropod intermediate hosts, and mammals; in turn, setting a foundation for establishing the environmental drivers of evolution for these parasites, and variable co-evolution among multiple taxonomic groups.
Our molecular analyses also revealed a dearth of genetic information for Hymenolepis tapeworms in universal data repositories. From all the species endemic to North America, there are only 2 with entries in the universal repositories of the NCBI (i.e., GenBank) for the homologous gene. This lack of representation may have contributed to the evident polytomy displayed in the phylogeny based on the mitochondrial locus. To rigorously document the existing diversity and functional significance of parasites across North America, it is critical that existing personal collections, new specimen accessions, and all associated host and molecular data are deposited as voucher representation within public research archives to enable future interpretation of their distributions, host associations, and evolutionary dynamics (Colella et al., 2021; Dunnum et al., 2017; Galbreath et al., 2019).
This practice is particularly important for Hymenolepis, a group that includes species of zoonotic importance, such as the cosmopolitan H. diminuta (Panti-May et al., 2020). Evidence gathered elsewhere suggests that there may be several species in the genus that have a zoonotic potential (Nkouawa et al., 2016), which may have been identified as H. diminuta based on the limited variability of characters available in the specimens used for the parasite diagnosis. In most cases these specimens include eggs and meristic characters as evidence for species level identification, yet there is overlap in the range of these measurements, such as the overall size of the embryophore and hooks. As demonstrated by Nkouawa et al. (2016), molecular screening allows a more precise identification of the specimens, which is not possible with a limited set of overlapping characters. Their results may suggest that some infections attributed to H. diminuta in humans may be induced by other species. As indicated by Panti-May et al. (2020) these identifications are based on the morphology of the eggs and not on the morphology of the adults. As seen in Table 3, the eggs feature very few meristic characters that overlap for several species.
This would be an unfortunate example of the “fallacy of the expected identification” (Hoberg & Soudachanh, 2021), one with important medical implications. In this phenomenon, scientists assign a name of a species that has been recorded in the host species elsewhere without the proper verification of the identity based on the analysis of the voucher specimen or DNA. Typically, this species is presumed common and widespread. We plea to the scientific community to properly voucher the material, which may allow other members of the community to evaluate it and offer a refined identification. An example of this practice includes the deposition of specimens initially identified as H. diminuta and collected from cotton rats (Faulkner & Lochmiller, 2000), examination of these specimens deposited in the USNM1382519 revealed that they belong to a different species. Another example includes the proper vouchering of specimens and the generation of barcodes accessible to the community (Sasaki et al., 2021). We propose that whenever possible, scientist excise tissues from specimens that will be eventually stained for identification and deposition in scientific collections.
Tapeworms are likely valuable indicators of biodiversity change across both geography and time, considering they require at least one arthropod intermediate host (Gardner & Campbell, 1992). Fluctuations in parasite abundance, distribution, and host association should prove important to our understanding of general biotic responses to anthropogenic environmental change.











 nueva página del texto (beta)
nueva página del texto (beta)


