Introduction
Type 2 diabetes (T2D) is a metabolic disorder characterized by hyperglycemia exceeding 110 mg/dL, often stemming from inadequate insulin production or insufficient insulin action [1]. One approach to managing hyperglycemia involves activating peroxisome proliferator-activated receptors of the gamma subtype (PPAR-γ) just like to synthetic 1,3-thiazolidine-2,4-diones. Natural PPAR-γ agonists include both saturated and unsaturated fatty acids, such as eicosanoids and prostaglandins. Conversely, synthetic ligands function as insulin-sensitizing drugs by either fully or partially activating PPAR-γ, thereby enhancing the expression of target genes crucial for glucose-sensing in pancreatic β-cells of diabetic individuals [2]. These target genes may include the solute carrier family 2 (facilitated glucose transporter), member 4 (GLUT-4), among others [2]. Another mechanism for regulating glucose levels involves the G protein-coupled receptor 40 (GPR40), predominantly found in pancreatic β-cells and enteroendocrine cells of the gut. Activation of GPR40 by medium to long chain fatty acids stimulates insulin secretion specifically when glucose concentration is high, without affecting insulin exocytosis during low glucose levels [3]. This intriguing mechanism hints at the potential of using small molecule agonists, such as 1,3-thiazolidine-2,4-diones, for GPR40 in treating T2D offering a novel approach as insulin secretagogues with a reduced risk of hypoglycemia [4].
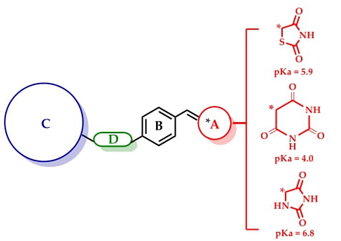
The aim of this study was to design, synthesize, and evaluate a series of ten 3-aryloxy-5-benzylidenebarbiturates and hydantoins as 1,3-thiazolidine-2,4-dione bioisosteres. These compounds were screened through in vitro, in vivo, and in silico methods, employing combined screening strategies. The objective was to develop a single molecule capable of targeting multiple pathways for treating diabetes, specifically aimed at activating PPAR-γ, GLUT-4, and GPR40. Multitarget efficacy aligns with the concept of polypharmacology, whereby multiple drug targets are modulated to achieve a desired therapeutic outcome [5]. Compounds 1-10 were designed by leveraging the structure of benzylidene-1,3-thiazolidine-2,4-dione compounds, which adhere to the characteristic 4-point unified pharmacophore seen in synthetic GPR40 and PPARγ agonists [2, 5, 6]. These features include: (A) an acidic group (1,3-thiazolidine-2,4-dione or surrogates); (B) a centrally substituted benzene; (C) a second lipophilic region; and (D) a flexible connector facilitating various conformations. Given these similarities, it is reasonable to infer that these barbiturates 1-5 (calculated pKa = 4.0) and hydantoins 6-10 (calculated pKa = 6.8) may exert similar agonistic effects on these targets, owing to their shared acidic properties with 1,3-thiazolidine-2,4-dione (calculated pKa = 5.9). Scheme 1 shows the reported antidiabetic unified pharmacophore of multitarget compounds [5]. It is important to note that the pharmacophore consists of four structural features starting with an acidic head that can be replaced by carboxylic acids or their surrogates that maintain an acidic pKa, such as the azaheterocycles mentioned above. The development of multi-target compounds for GPR40, PPAR-γ and some other proteins implied in the diabetic pathogenesis may provide additional therapeutic benefits in preventing or delaying the development of diabetic complications.
Experimental
Chemistry
Reagents, materials, and solvents sourced from Merck/Sigma-Aldrich were utilized without further purification. Melting points, left uncorrected, were determined using an EZ-Melt MPA120 automated melting point apparatus. Reaction progress was monitored via TLC on 0.2 mm precoated silica gel 60 F254 Merck plates. NMR spectra were acquired on a Varian Oxford instrument operating at 600 MHz for 1H nuclei and 150 MHz for 13C nuclei. Chemical shifts (δH and δC values) are reported in parts per million (ppm), while homocoupling patterns are expressed in Hertz (Hz). Mass spectrometry (EI-MS) was conducted on a JEOL JMS-700 spectrometer using the electronic impact method.
General procedure for the synthesis of barbituric derivatives 1-5
A mixture of barbituric acid (1 equivalent) and piperidine (30 % mol) were dissolved in 10 mL of toluene and stirred at 60 °C for 20 min. Besides, 1.1 equivalents of ether-aldehyde precursors 10-15 and benzoic acid (30 % mol) were added and heated to reflux (~90°C) for 2-10 h. Continuous removal of water formed was allowed with the help of the Dean-Stark apparatus. The reaction mixture was cooled, and the yellow solid was filtered off and dried.
5-(3-methoxy-4-(quinolin-2-ylmethoxy)benzylidene)pyrimidine-2,4,6 (1H,3H,5H)-trione (1). Orange crystals, yield 93 %, m.p. 187 °C (dec). 1H NMR (600 MHz, DMSO d-6) δ: 2.79 (s, 3H), 5.02 (s, 2H), 6.72 (dd, 1H, J o = 7.67 Hz), 6.79 (d, 1H, J o = 8.32 Hz), 6.88 (dd, 1H, J o = 7.45 Hz), 7.08 (m, 3H), 7.38 (m, 1H), 7.51 (s, 1H), 10.24 (s, 1H), 10.38 (s,1H), ppm. 13C NMR (150 MHz, DMSO d-6) δ: 56.0 (C-7), 72.2 (C-8), 112.2 (C-2), 113.8 (C-5), 119.3 (C-10), 120 (C-3’), 126.9 (C-1), 127.63 (C-6’), 128.3 (C-6), 128.9 (C-5’), 129.7 (C-8’), 130.2 (C-7’), 133.9 (C-8a’), 139.0 (C-4’), 145.5 (C-3, C-4), 147.3 (C-13), 148.8 (C-11), 151.1 (C-9), 158.2 (C-2’), 168.2 (C-12), ppm. MS: m/z (% rel. int.) 403 (M+, 1 %), 91 (M-312, 100 %), 148 (M-255, 45 %).
5-(3-methoxy-4-(naphthalen-1-ylmethoxy)benzylidene)pyrimidine-2,4,6 (1H,3H,5H)-trione (2). Yellow powder, yield 86 %, m.p. 202.3 - 204.2°C. 1H NMR (600 MHz, DMSO d-6) δ: 2.93 (s, 3H), 4.84 (s, 2H), 6.56 (d, 1H, J o = 8.56 Hz), 6.73 (m, 3H), 6.85 (d, 1H, J o = 6.96 Hz), 7.08 (dd, 1H, J m = 2.04 Hz, J o = 6.8 Hz), 7.12 (d, 1H, J o = 8.34 Hz), 7.16 (d, 1H, J o = 6.78 Hz), 7.24 (m, 1H), 7.43 (s,1H), 7.57 (d, 1H, J m = 2.1 Hz), 10.34 (s, 1H), 10.47 (s, 1H), ppm. 13C NMR (150 MHz, DMSO d-6) δ: 55.9 (C-7), 68.9 (C-8), 112.9 (C-2), 115.9 (C-5), 117.5 (C-10), 124.3 (C-3'), 125.8 (C-6'), 126.0 (C-6), 126.5 (C-7'), 127 (C-1), 127.4 (C-2'), 128.9 (C-4'), 129.4 (C-5'), 131.6 (C-8'), 131.8 (C-1'), 132.2 (C-8a'),133.7 (C-4a'), 148.4 (C-4), 150.6 (C-3), 153.1 (C-9), 155.8 (C-13), 162.7 (C-11),164.4 (C-12), ppm. MS: m/z (% rel. int.) 402 (M+, 1 %) 128 (M-274, 100 %) 141 (M-261, 25 %).
5-(4-([1,1'-biphenyl]-3-ylmethoxy)-3-methoxybenzylidene)pyrimidine-2,4,6(1H,3H,5H)-trione (3). Yellow powder, yield 83 %, m.p. 173.2 - 175.4 °C. 1H NMR (600 MHz, DMSO d-6) δ: 3.61 (s, 3H), 5.03 (s, 2H), 6.62 (s, 2H), 6.84 (d,1H, J o = 7.62 Hz), 7.34 (t, 1H, J o = 7.38 Hz), 7.39 (d, 1H, J o = 7.56 Hz), 7.41 (d, 1H, J o = 7.32 Hz), 7.45 (t, 3H, J o = 7.62 Hz), 7.58 (d, 1H, J o = 7.56 Hz), 7.63 (d, 2H, J o = 7.56 Hz), 7.69 (s, 1H), 9.94 (s, 2H), ppm. 13C NMR (150 MHz, DMSO d-6) δ: 55.9 (C-7), 70.7 (C-8), 112.2 (C-2), 113.8 (C-5), 119.3 (C-10), 126.5 (C-4'), 126.5 (C-2'), 127.1 (C-2'', C-6''), 127.2 (C-6'), 127.9 (C-3'), 129.4 (C-3'', C5''), 129.5 (C-4''), 138.7 (C-9), 140.4 (C-4), 140.6 (C-3), 145.7 (C-6), 148.8 (C-12), 151.1 (C-11, C-13), ppm. MS: m/z (% rel. int.) 428 (M+, 1 %), 167 (M-261, 50 %).
4'-((2-methoxy-4-((2,4,6-trioxotetrahydropyrimidin-5(2H)-ylidene)methyl) phenoxy)methyl)-[1,1'-biphenyl]-2-carbonitrile (4). Yellow powder, yield 79 %, m.p. 239.9 - 240.0 °C. 1H NMR (600 MHz, DMSO d-6) δ: 3.33 (s, 3H), 4.83 (s, 2H), 7.08 (s, 1H), 7.12 (m, 4H), 7.29 (m, 1H), 7.40 (d, 1H, J o = 8.89 Hz), 7.41 (dd, 1H, J o = 8.52 Hz), 7.45 (t, 1H J o = 8.02 Hz), 7.76 (d, 1H, J o = 8.89 Hz), 7.79 (s, 1H), 7.92 (d, 1H, J m =1.49 Hz), 10.68 (s, 1H), 10.8 (s, 1H), ppm.13C NMR (150 MHz, DMSO d-6): δ: 56.3 (C-7), 70.3 (C-8), 110.9 (C-6''), 113.1 (C-2), 116.1 (C-5), 117.9 (CN), 119.3 (C-10), 126.0 (C-6), 128.7 (C-1), 129 (C-3', C-5'), 129.7 (C-2', C-6'), 130.9 (C-2''), 132.1 (C-4''), 134.3 (C-3''), 134.6 (C-5''), 137.7 (C-1'), 138.4 (C-4'), 144.9 (C-1''), 148.7 (C-4), 150.9 (C-3), 153.3 (C-9), 156.1 (C-13), 163.1 (C-11), 164.7 (C-12), ppm. MS: m/z (% rel. int.) 453 (M+, 1 %) 192 (M-261, 100 %), 165 (M-288, 10 %).
5-(3-methoxy-4-(2-morpholinoethoxy)benzylidene)pyrimidine-2,4,6(1H,3H,5H)-trione (5). Beige crystals, yield 56 %, m.p. 218.3 - 221.9 °C. 1H NMR (600 MHz, DMSO d-6) δ: 3.42 (m, 10H), 3.72 (s, 3H), 4.12 (s, 2H), 7.49 (dd, 1H Jo = 7.10 Hz), 7.61(dd, 1H, J o = 7.16 Hz), 7.94 (d, 1H, J o = 7.62 Hz), 8.44 (s, 1H), 11.09 (s, 2H), ppm.13C NMR (150 MHz, DMSO d-6) δ: 43.7(C-7), 52.66 (C-9), 55.5 (C-2', C-3'), 56.1 (C-1', C-4'), 64.5 (C-8), 111.7 (C-2), 119.0 (C-11), 128.6 (C-5), 129.2 (C-6), 132.9 (C1), 144.7 (C-3), 148.6 (C-4), 150.7 (C-10), 151.6 (C-12, C-14), 167.7 (C-13), ppm. MS: m/z (% rel. int.) 375 (M+, 1 %), 128 (M-247, 100 %), 85 (M-290, 20 %).
General procedure for the synthesis of hydantoin derivatives 6-10
A mixture of hydantoin (1 equivalent) and piperidine (60 % mol) were dissolved in 10 mL of toluene and stirred at 40 °C for 20 min. On the other hand, 1.1 equivalents of ether-aldehyde precursors 12-16 and benzoic acid (60 % mol) were added and heated to reflux (~90°C) for 10-15 h. Continuous removal of water formed was allowed with the help of the Dean-Stark apparatus. The reaction mixture was cooled, and the solid was filtered off and dried.
(Z)-5-(3-methoxy-4-(quinolin-2-ylmethoxy)benzylidene)imidazolidine-2,4-dione (6). Green powder, yield 62 %, m.p. 245.1 - 247.0 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.90 (s, 1H), 5.67 (s, 1H), 6.50 (s, 1H), 7.26 (d, 1H, J m = 1.98 Hz), 7.33 (t, 2H), 7.63 (t, 1H, J o = 7.63 Hz), 7.68 (ddd, 1H, J o = 7.44 Hz, J m = 1.98 Hz), 7.77 (d, 1H, J o = 7.02 Hz), 8.05 (d, 1H, J o = 8.28 Hz), 8.09 (dd, 1H, J m = 1.86 Hz, J o = 6.6 Hz), 8.21 (d, 1H, J o = 7.98 Hz), 10.66 (s, 1H), 11.27 (s, 1H), ppm. 13C NMR (150 MHz, DMSO-d6) δ: 56.1 (C-7), 68.8 (C-8), 109.7 (C-9), 113.4 (C-2),125.7 (C-6), 126.3 (C-5), 126.5 (C-3’), 126.7 (C-1), 127.1 (C-6’), 128.8 (C-10), 129.0 (C-4a’), 129.3 (C-5’), 131.6 (C-8’), 132.8 (C-7’), 133.7 (C-4’), 148.8 (C-4), 149.6 (C-3), 156.2 (C-8a’), 158.8 (C-12), 166.1 (C-2’), 174.3 (C-11), ppm; m/z (% rel. int) 375 (M+, 1 %), 264.2 (M-111, 36 %), 222 (M-153, 12 %).
(Z)-5-(3-methoxy-4-(naphthalen-1-ylmethoxy)benzylidene)imidazolidine-2,4-dione (7). Yellow powder, yield 66 %, m.p. 233.1 - 235.2 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.78 (s, 3H), 5.54 (s, 2H), 6.38 (s, 1H), 7.14 (d, 1H, J m = 1.98 Hz), 7.20 (dd, 1H, J m = 1.92 Hz, J o = 8.76 Hz), 7.21(d, 1H, J o = 8.4 Hz), 7.50 (t, 1H, J o = 7.74 Hz), 7.55 (m, 2H), 7.65 (d, 1H, J o = 7.86 Hz), 7.92 (d, 1H, J o = 6.9 Hz), 7.96 (d, 1H, J o = 7.68 Hz), 8.09 (d, 1H, J o = 8.76 Hz), 10.49 (s,1H), 11.16 (s, 1H), ppm. 13C NMR (150 MHz, DMSO-d6) δ: 56.1 (C-7), 68.8 (C-8), 109.7 (C-9), 113.4, (C-2), 114.1 (C-6), 123.3 (C-5), 125.7 (C-3'), 126.3 (C-6' y C-7'), 126.5 (C-2'), 126.7 (C-4'), 128.8 (C-5'), 129.0 (C-1), 129.3 (C-10), 131.6 (C-8'), 132.8 (C-4a'), 148.8 (C-1' y C-8a'), 156.2 (C-4),158.8 (C-3), 166.1 (C-12), 174.3 (C-11), ppm; m/z (% rel. int) 374 (M+, 1 %), 142 (M-232, 100 %), 115.0 (M-259, 30 %).
(Z)-5-(4-([1,1'-biphenyl]-3-ylmethoxy)-3-methoxybenzylidene)imidazolidine-2,4-dione (8). Yellow powder, yield 84 %, m.p. 238.9 - 241.2 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.82 (s, 3H), 5.19 (s, 2H), 6.36 (s, 1H), 7.07 (d, 1H, J o = 8.22 Hz), 7.14 (s, 1H), 7.17 (dd, 1H, J m = 2.04 Hz, J o = 7.86 Hz), 7.32-7.37 (m, 1H), 7.41-7.48 (m, 4H), 7.62 (d, 2H, J o = 7.68 Hz), 7.67 (s, 1H), 7.72 (s, 1H), 9.82 (s, 1H),10.62 (s, 1H), ppm. 13C NMR (150 MHz, DMSO-d6) δ: 57.3 (C-7), 65.8 (C-8), 119.1 (C-9), 123.0 (C-2), 123.7 (C-5), 130.6 (C-10), 132.9 (C-6), 136.2 (C-6’), 136.3 (C-2’), 136.4 (C-1), 136.7 (C-5’), 136.9 (C-3’’ y C-5’’), 137.6 (C-4’),139.0 (C-2’’), 139.2 (C-4’’y C-6’’), 147.7 (C-1’), 150.0 (C-1’’), 150.4 (C-3’), 158.3 (C-4), 159.2 (C-3), 168.4 (C-12), 183.9 (C-11), ppm; m/z (% rel. int) 400 (M+, 2 %), 121 (M-279, 45 %), 93 (M-307, 56 %).
(Z)-4'-((4-((2,5-dioxoimidazolidin-4-ylidene)methyl)-2-methoxyphenoxy) methyl)[1,1'-biphenyl]-2-carbonitrile (9). Yellow powder, yield 76 %, m.p. 226.1 - 228.0 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.84 (s, 3H), 5.20 (s, 2H), 6.37 (s, 1H), 7.09 (d, 1H, J o = 8.4 Hz), 7.15 (d, 1H, J m = 2.04 Hz), 7.18 (d,1H, J m = 2.04 Hz, J o = 8.34 Hz), 7.59 (m, 6H), 7.78 (dd, 1H, J o = 7.7 Hz, J m = 2.6 Hz), 7.94 (d, 1H, J o = 7.8 Hz), 10.48 (s,1H), 11.15 (s, 1H), ppm. 13C NMR (150 MHz, DMSO-d6) δ: 56.2 (C-7), 69.8 (C-8), 109.5 (C-6''), 110.6 (C-2), 113.3 (C-9), 113.8 (C-5), 119.0 (CN) 123.3 (C-6 y C-10), 126.5 (C2''), 126.7 (C-2', C-6'), 128.5 (C-4''), 128.7 (C-1), 129.2 (C-3''),130.6 (C-5''), 134.0 (C-5'), 134.3 (C-3'), 137.8 (C-1'),138.0 (C-4'), 144.6 (C-1''), 148.7 (C-4), 149.6 (C-3), 156.2 (C-12), 166.1 (C-11), ppm; m/z (% rel. int) 425 (M+, 1 %), 281.0 (M-144, 40%), 192 (M-233, 100 %).
(Z)-5-(3-methoxy-4-(2-morpholinoethoxy)benzylidene)imidazolidine-2,4-dione (10). Yellow powder, yield 84 %, m.p. 237.3 - 238.9 °C. 1H NMR (600 MHz, DMSO-d6) δ: 2.69 (t, 2H, J o= 5.64 Hz), 3.57 (t, 4H, J o = 4.44 Hz), 3.82 (s, 3H), 3.84 (s, 4H), 4.10 (t, 2H, J o = 5.84 Hz), 6.38 (s,1H), 6.99 (d,1H, J o = 8.28 Hz), 7.12 (d, 1H, J m = 1.92 Hz), 7.17(dd, 1H, J m = 1.93 Hz, J o = 9.06 Hz), 10.48 (s, 1H), 10.61 (s, 1H), ppm. 13C NMR (150 MHz, DMSO-d6) δ: 47.7 (C-9), 54.1 (C-7), 56.2 (C-1’, C-4’), 57.4 (C-2’ , C-3’), 66.7 (C-8), 109.6 (C-10), 113.3 (C-3), 113.6 (C-2), 123.4 (C-6), 126.3 (C-1), 126.7 (C-11), 149.0 (C-4), 149.4 (C-3), 166.1 (C-13), 174.3 (C-12), ppm; m/z (% rel. int) 347 (M+, 1 %), 296 (M-51, 9 %), 264 (M-83, 32 %).
General method of synthesis for precursors 12 - 16.
A solution of 4-hydroxy-3-methoxybenzaldehyde (11) and potassium carbonate (2.2 equivalents) was prepared in acetonitrile and stirred at room temperature for thirty minutes. Aryl bromides 17-21 (1.1 equivalents) were added gradually, and the mixture was heated to reflux for 2-5 h. Upon completion of the reaction, the solvent was evaporated under reduced pressure, and the resulting solids were washed with cold water to remove excess K2CO3. The crude solid products underwent recrystallization in ethanol.
3-methoxy-4-(quinolin-2-yl-methoxy)benzaldehyde (12). Beige crystals, yield 52 %, m.p. 108.3 - 109.9 °C. 1H NMR (600 MHz, DMSO-d6) δ: 4.00 (s, 3H), 5.55 (s, 2H), 7.05 (d, 1H, J o = 8.22 Hz), 7.37 (dd, 1H, J m = 1.75 Hz, J o = 7.02 Hz), 7.46 (d, 1H, J m = 1.7 Hz), 7.56 (dd, 1H, J m = 1.4 Hz, J o = 7.8 Hz), 7.68 (d, 1H, J o = 8.5 Hz), 7.75 (dd, 1H, J m = 2.5 Hz, J o = 7.7 Hz), 7.84 (d, 1H, J o = 8.1 Hz), 8.19 (d, 1H, J o = 4.98 Hz), 8.9 (d, 1H, J o = 5.38 Hz), 9.83 (s,1H), ppm; m/z (% rel. int) 293 (M+, 25 %), 142 (M-151, 100 %)
3-methoxy-4-(naphthalen-1-ylmethoxy)benzaldehyde (13). White crystals, yield 95 %, m.p. 98.5 - 99.8 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.80 (s, 3H), 5.66 (s, 2H), 6.60 (d, 1H, J o = 5.64 Hz), 7.43 (d, 1H, J m = 2.0 Hz), 7.48 (d, 1H, J o = 8.28 Hz), 7.53 (dd, 1H, J m =1.32 Hz, J o = 7.02 Hz), 7.56 (m, 2H), 7.69 (d, 1H, J o = 6.96 Hz), 7.95 (d, 1H, J o = 8.28 Hz), 7.99 (d, 1H, J o = 7.38 Hz), 8.10 (d, 1H, J o = 7.86), 9.87 (s, 1H), ppm; m/z (% rel. int) 292 (M+, 1 %), 264 (M-28, 100 %) 128 (M-164, 100 %)
4-([1,1'-biphenyl]-3-ylmethoxy)-3-methoxybenzaldehyde (14). White crystals, yield 64 %, m.p. 84.1 - 85.4 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.84 (s, 3H), 5.30 (s, 2H), 7.32 (d, 1H, J o = 8.52 Hz), 7.38 (t, 1H, J o = 6.78 Hz), 7.43 (s, 1H), 7.48 (m, 4H),7.56 (d, 1H, J o = 8.10 Hz), 7.66 (m, 3H), 7.77 (s, 1H), 9.85 (s, 1H), ppm; m/z (% rel. int) 318 (M+, 12.5 %) 167 (M-151, 100 %).
4'-((4-formyl-2-methoxyphenoxy)methyl)-[1,1'-biphenyl]-2-carbonitrile (15). White crystals, yield 64 %, m.p. 161.0 - 162.3 °C. 1H NMR (600 MHz, DMSO-d6) δ: 3.86 (s, 3H), 5.31 (s, 2H), 7.32 (d, 1H, J o = 8.22 Hz), 7.44 (d, 1H, J m = 1.92 Hz), 7.56 (dd, 1H, J m = 1.98 Hz, J o = 8.4 Hz), 7.59 (dd, 1H, J m = 2.82 Hz, J o = 7.86 Hz), 7.63 (m, 5H), 7.80 (dd, 1H, J m = 3.36 Hz, J o = 7.45 Hz), 7.97 (d, 1H, J o =7.44 Hz), 9.86 (s,1H), ppm; m/z (% rel. int) 343 (M+, 5 %) 192 (M-151, 100 %).
3-methoxy-4-(2-morpholinoethoxy)benzaldehyde (16). White crystals, yield 84 %, m.p. 268.7 - 269.9 °C. 1H NMR (600 MHz, DMSO-d6) δ: 2.43 (t, 4H), 3.27 (t, 4H), 3.32 (s, 3H), 3.50 (t, 2H), 4.28 (t, 2H), 6.96 (d, 1H, J o = 8.28 Hz), 7.15 (d, 1H, J m = 1.92 Hz), 7.32 (dd, 1H, J o = 9.06 Hz, J m = 1.92 Hz), 8.88 (s,1H), ppm; m/z (% rel. int) 265 (M+, 1%), 236 (M-29, 5 %), 98 (M-167, 100 %).
Biological Assays
Gene expression of GLUT-4 and PPAR-γ
C2C12 myocytes (ATCC/CRL-1772) were incubated and maintained in DMEM medium supplemented with 10 % fetal bovine serum (SFB), 0.5 mM sodium pyruvate, 1 mM L-glutamine, 0.05 mM non-essential amino acids, and 0.1 mg/L gentamicin, under a humidified atmosphere of 5 % CO2 at 37 °C [3]. The cells were treated with a varying concentrations of compounds 1, 2 and 4 by 24 h to assess mRNA expression levels of GLUT-4 and PPAR-γ.
Cell culture of the RIN-m5F line
RIN-m5F pancreatic insulinoma cells (ATCC/CRL-11605) were procured and cultured in 75 cm2 flasks until reaching confluence. The cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (SFB), 0.5 mM sodium pyruvate, 1 mM L-glutamine, 0.05 mM non-essential amino acids, and 0.1 mg/L gentamicin, under a humidified atmosphere of 5% CO2 at 37°C [3]. The culture medium was refreshed every 48 h.
Evaluation of the relative expression levels of PPAR-γ, GLUT4, and GPR40
C2C12 and RIN-m5F cells were plated individually in 6-well plates at a density of 8x104 cells per well and treated with compounds 1, 2, and 4 at a concentration of 1 µM. Muscle cells were treated with 5 µM pioglitazone as a control, while β-pancreatic cells received 400 µM glibenclamide. Following a 24-h incubation period, total RNA was extracted from the cells using TRIZOL reagent. The RNA samples were assessed for purity by measuring absorbance at 260 and 280 nm, with an OD ratio (260/280) of 1.9 ± 0.2, indicating minimal contamination. Reverse transcription of 2 μg mRNA was carried out using the ImProm II reverse transcription system (Promega, Wisconsin, USA). The resulting cDNA was subjected to amplification using SYBR Green master mix (ThermoScientific, USA) and primers targeting the PPAR-γ, GLUT4, and GPR40 genes, with 36B4 and β-actin serving as reference genes. Amplification was performed using a rotor-gene system (Techne, PrimePro48, UK). ΔCt values were computed for each sample and gene of interest by subtracting the Ct value of the reference gene from the Ct value of the target gene. Relative changes in gene expression (ΔΔCt) were determined by subtracting the ΔCt of the control group from the ΔCt of the test group, and then reporting the result as 2−ΔΔCt [2,3,7,8].
Insulin secretion
RIN-m5F cells were plated in 6-well plates at a density of 8 x 104 cells per well and allowed to reach 70-80% confluence. Following this, the cells were treated with glibenclamide (400 µM) and compounds 1, 2, and 4 (1 µM) for 24 h [9].
In vivo antidiabetic assay
Animals
Male C57BL/6 mice, weighing 25 ± 5 g, were accommodated in animal cages with a 12-h light-dark cycle. The mice were kept in a controlled environment at 25 °C, with free access to water and food. All experiments involving mice were conducted in accordance with protocols approved by the Mexican government NOM-065-ZOO-1999 and NOM-033-ZOO-2014 and were further authorized by the Institutional Ethics Committee of the Universidad Autónoma Metropolitana (dictum 1857), adhering to the guidelines outlined in the US National Institutes of Health Publication #85-23, revised in 1985.
Oral glucose tolerance test
Normoglycemic mice were separated in five groups (n = 6):
Group 1: Compound 1, 100 mg/kg.
Group 2: Compound 2, 100 mg/kg.
Group 3: Compound 4, 100 mg/kg.
Group 4: control (isotonic saline solution, ISS).
Group 5: positive control (glibenclamide, pioglitazone or linagliptin, 5 mg/kg).
A load of 2 g/kg of glucose or sucrose solution was administered to mice 30 min after test samples. Then blood samples were obtained at time 0 (before oral administration), 0.5, 1, 1.5, 2, and 3 h after the vehicle, positive control, and compounds administrations, from the caudal vein. Glycemia was estimated by the glucose dehydrogenase method using a commercial glucometer (Accu-Chek, Performa; Roche®). The percentage change of glycemia for each group was calculated in relation to the initial (0 h) level [10].
Statistical analysis
To examine the disparities in the percentage variation of glycemia and the quantification of in vitro mRNA PPAR-γ, GLUT-4, and GPR40 expression, we utilized ANOVA, supplemented with a Dunnett's multiple test. All data are presented as mean ± S.E.M. with significance set at p < 0.05, and the analysis was conducted using GraphPad Prism 5.0.
Molecular docking
Docking calculations were conducted using the licensed Molecular Operating Environment (MOE) version 2020.0901 [11]. The crystal structure of PPAR-γ complexed with rosiglitazone PDB ID: 4EMA, and 4PHU complexed with TAK-875 at a resolution of 2.54 and 2.33 Å respectively, were retrieved from the Protein Data Bank (http://www.rcsb.org/pdb) [12, 13]. The unnecessary molecules were removed, the hydrogen atoms and charges were adjusted using the Amber14:EHT forcefield from the MOE suite (Chemical Computing Group Inc. http://www.chemcomp.com). The 3D structure of compounds 1, 2 and 4 was constructed and minimized in MOE using the MMF94xforcefield. Docking simulations were conducted, considering all residues within a 4.5 Å sphere centered on the atoms of the cocrystallized ligand. The Triangle Matcher and Alpha Triangle was selected as the placement function, the scores were calculated with the London ΔG function and the selection of the best poses was made using the GBVI/WSA ΔG (Generalized-Born Volume Integral/Weighted Surface area). This process was validated by reproducing, through docking, the same pose as the cocrystallized ligand in the PPAR-γ (RMSD = 1.0369 Å) with a score of -8.4423 Kcal/mol and GPR40 crystal structure with a score of -11.3657 Kcal/mol (RMSD = 0.4234 Å). A value less than 2 Å suggests that the docking simulation parameters are effective in accurately reproducing the ligand pose within the protein. One hundred conformations were generated for each ligand, and the top-ranked conformation, determined by the docking score, was chosen for subsequent investigations in molecular docking. Following the molecular docking process, we scrutinized the optimally calculated binding poses, and graphical representations were generated using Surface Maps and Ligand Interaction tools in MOE and The PyMOL Molecular Graphics System, Version 3.0, Schrödinger, LLC. [14].
In silico prediction of biopharmaceutics, pharmacokinetics, and toxicological profile
For the estimation of the ADMET properties, to build the in silico pharmacological consensus analysis [15], we employed online programs like Molinspiration (https://www.molinspiration.com) [16], ADMETLab 3.0 (https://admetlab3.scbdd.com/) [17], MetaTox 2.0 (https://www.way2drug.com/metatox) [18]. The operation of these ADMET predictors involves three main steps: A) Input: cleaning individual molecules or batches of molecules using SMILES notation to input chemical structures; B) Operation: calculating the ADMET properties of these molecules using deep learning models; and C) Output: determining which result files to return. The modular design of this system allows for flexible combinations of functionalities. The calculation of ADMET properties includes molecular basic properties, physical chemistry, medicinal chemistry, absorption, distribution, metabolism, excretion, and toxicity, among others.
Results and discussion
Chemistry
Compounds 1-10 were prepared starting from 4-hydroxy-3-methoxybenzaldehyde (11), which was reacted via SN2 with adequately substituted arylbromides 17-21, to obtain the ether-aldehyde precursors 12-16. These compounds were then condensed under Knoevenagel conditions with barbituric acid to afford compounds 1-5 or with 2,4-imidazolidinedione (hydantoin) to give compounds 6-10 (Scheme 2).
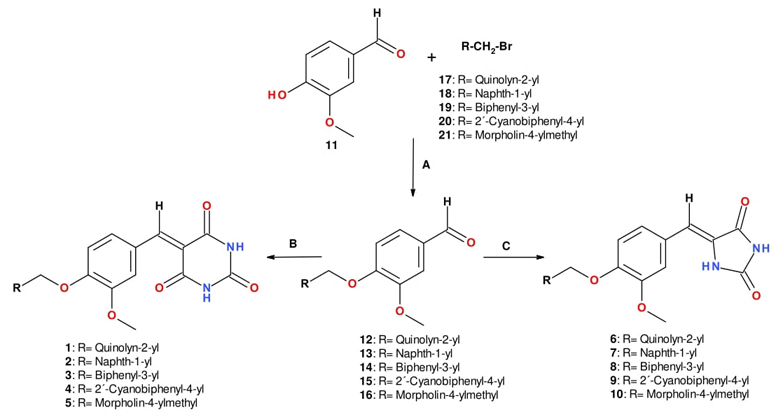
Scheme 2 Synthesis of compounds 1-10. Reagents and conditions: (A) K2CO3, acetonitrile, reflux; (B) Barbituric acid, piperidine (0.3 equiv), benzoic acid (0.3 equiv), toluene, Dean-Stark apparatus, reflux; (C) 1,3-imidazolidine-2,4-dione (hydantoin), piperidine (0.6 equiv), benzoic acid (0.6 equiv), toluene, Dean-Stark apparatus, reflux.
To synthesize the hydantoin series, it was imperative to double the quantity of the Knoevenagel additives (piperidine and benzoic acid) owing to the low yields obtained from the initial procedure. This could be attributed to the poor solubility of hydantoin. When hydantoin reacts with piperidine in an acid-base reaction, it forms a more soluble anion. This process requires twice the amount of both additives in the Knoevenagel reaction.
In silico Pharmacological Consensus Analysis
We conducted an in silico pharmacological consensus analysis (PHACA) and summarized the results in Table 1 using a traffic light system [15]. PHACA integrates calculations from biopharmaceutical properties, pharmacodynamics and pharmacokinetic predictions, toxicity assessments, and additional experimental data. The rationale behind pharmacological consensus analysis lies in the agreement among multiple predicted parameters indicating a compound's activity, low toxicity, and favorable pharmacokinetic profile. Accordingly, selecting a compound with a high score across various predictions enhances confidence in its suitability for synthesis. Thus, a compound receiving high scores from multiple predictions is more likely to exhibit desirable behavior in biological assays compared to one with high scores from a single prediction [15].
Table 1 presents the results of Pharmacological Consensus Analysis, which involves assessing various properties and assigning them a color-coded classification. Unsatisfactory profiles are marked in red, satisfactory profiles in yellow, and very satisfactory profiles in green. A final score is computed by summing the numeric values assigned to each profile (very satisfactory: +1; satisfactory: 0; unsatisfactory: -1). A higher score indicates superior combined pharmaceutical properties, prioritizing the molecule for synthesis and/or experimental biological evaluation. The analysis utilizes validated online programs like Molinspiration (Rule of 5) [16], ADMETLab (Human intestinal absorption Ames toxicity, carcinogenicity) [17], MetaTox 2.0 (molecular targets) [18], and ACD ToxSuite (hERG blockade, CYP inhibition) [19]. The biosimulation results suggest that compounds 1-10 demonstrate safety and favorable pharmacokinetic (PK) and pharmacodynamic (PD) properties, enhancing both permeability and intestinal absorption.
Table 1 In silico pharmacological consensus analysis (PHACA).
| Barbiturates | Hydantoins | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Pharmacodynamics | Molecular Docking | ||||||||||
| Molecular targets (GPR40, PPAR-γ) | |||||||||||
| Pharmacokinetics | Human intestinal absorption | ||||||||||
| PGP inhibition | |||||||||||
| Toxicity | CYP´s inhibition | ||||||||||
| hERG blockage | |||||||||||
| Ames toxicity | |||||||||||
| Carcinogenic | |||||||||||
| OECD/LD50 | |||||||||||
| Biopharmaceutics Properties | Solubility | ||||||||||
| Permeability | |||||||||||
| Final Score | 10 | 10 | 8 | 10 | 5 | 5 | 7 | 6 | 6 | 5 | |
Moreover, the compounds demonstrate potential in silico affinity for GPR40 and PPAR-γ and display favorable predictions for low cardiotoxicity, with no indications of carcinogenic or mutagenic effects. These descriptors are crucial in drug design to anticipate appropriate biopharmaceutical and pharmacokinetic profiles. Based on the analysis scores, compounds 1, 2, and 4 present the most promising biosimulation profiles with the highest score values (Final Score =10) (Table 1) and were thus selected as safe computational hits for prioritized in vitro and in vivo assays. This was the cut-off value (Final Score =10) that was taken into account to prioritize, using PHACA, the molecules that would be candidates for bioassays. On the other hand, for the hydantoin series (6-10), all resulted in low scores below 7, and compound 3 had a score of 8, indicating they were not prioritized for further in vitro or in vivo experiments to save financial resources and reduce the number of animals used.
In vitro results
Relative expression of PPARγ and GLUT-4
Myocytes are cells with active metabolic like to adipocytes, in both types cells it’s possible develop the insulin resistance. In this paper, myocytes were used to assess the impact of selected safe computational hits 1, 2 and 4 on PPAR-γ and GLUT-4 expression. Cells were treated with selected compounds, and pioglitazone (5 μM) as a positive control, for 24 h, and mRNA expression levels were evaluated. Fig. 1 illustrates that only compounds 1 and 2 significantly elevate relative expression levels of PPAR-γ (approximately three to fourfold). Activation of PPAR-γ has the potential to reduce glucose levels in diabetic individuals by mitigating insulin resistance. Furthermore, our findings suggest that compound 1 induces GLUT-4 expression more effectively than pioglitazone. Multiple lines of evidence suggest that elevated levels of GLUT-4 expression in skeletal muscle play a crucial role in regulating glucose homeostasis [2,3,6].
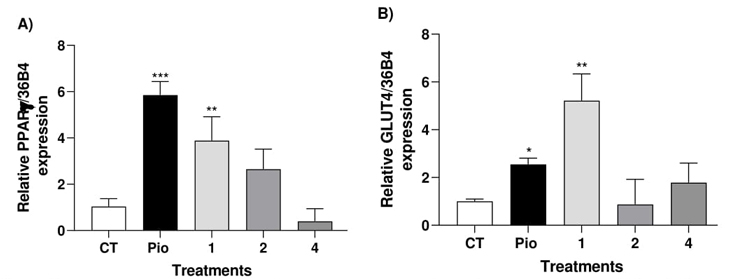
Fig. 1 Effect of compound 1, 2, 4 and Pioglitazone (Pio) on relative PPAR-γ expression (A) and GLUT-4 levels (B) in C2C12 myocytes. Results are expressed as relative expression of mRNA (mean ± S.E.M, n = 4). *p < 0.05 compared with control.
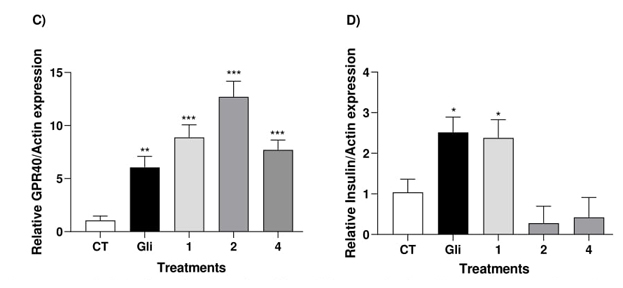
Fig. 2 Effect of compound 1, 2, 4 and glibenclamide (Gli) on relative GPR40 expression (C) and insulin levels (D) in RInm5F cells. *p < 0.05 compared with control.
Conversely, compounds 1, 2, and 4 exhibited a notable 7- to 12-fold increase in GPR40 expression (Fig. 2). To confirm whether the substantial increase in GPR40 expression induced insulin release, we assessed the compounds in RINm5F cells. These cells are involved in insulin secretion and intracellular calcium release [3]. We observed a reasonable insulin secretion induced by compound 1, comparable to that induced by glibenclamide.
In vivo antidiabetic effect of compounds 1, 2 and 4
Compounds 1, 2 and 4 were the most active against three targets identified as critical elements in diabetes in this work. Consequently, they were selected to evaluate their in vivo activity in a glucose tolerance test. Glibenclamide, pioglitazone and linagliptin served as positive controls for antidiabetic activity. The effects of compounds 1, 2, and 4 were assessed following a single oral dose of 100 mg/kg administered via the intragastric route (Fig. 3).
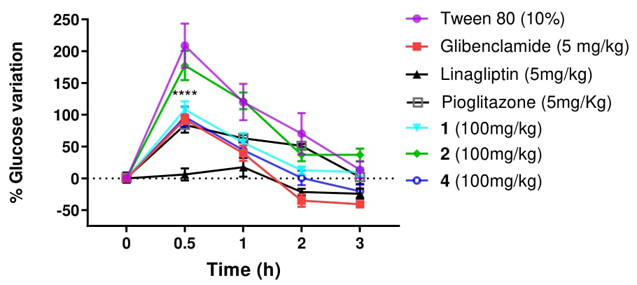
Fig. 3 In vivo oral Glucose Tolerance Test: Assessment of the impact of compounds 1, 2, and 4 on blood glucose levels following a single oral dose of 2 g/kg glucose in normoglycemic male C57BL/6 mice. Each plot displays the mean values along with the standard error of the mean (SEM) for six separate experiments. ***p < 0.05 indicates significance compared to the control group.
In the glucose tolerance test conducted with C57BL/6 mice (Fig. 3), the efficacy of compounds 1 and 4 is evident, as they demonstrate effectiveness within the initial 30 min of the test, mitigating the hyperglycemic peak. Furthermore, a notable decrease in glucose concentration is observed over the subsequent 60 min, with significant differences compared to vehicle. Compound 4 maintains its decreasing trend throughout the experiment, reaching levels below the basal levels along with glibenclamide, linagliptin and pioglitazone (positive controls), while compound 1 remains relatively stable.
Compound 1 demonstrated a clear increase in relative PPAR-γ expression, which was consistent with a rise in GLUT4 levels. A similar pattern was observed with the increase in GPR40 and insulin secretion. Conversely, compound 4 exhibited poor in vitro activity but significantly increased GPR40 expression by more than sevenfold, which may have contributed to its notable in vivo antihyperglycemic effect. This could be attributed to the reactive nitrile group in compound 4, which can form reversible covalent adducts with proteins, primarily through reactive cysteine or serine side chain residues. One potential additional target implicated in this reversible covalent effect is dipeptidyl peptidase-4 (DPP-4), a serine protease that inactivates incretin hormones and is a widely exploited target for treating type 2 diabetes mellitus. It has been established that the nitrile warhead in vildagliptin and saxagliptin forms a reversible covalent bond with the Ser-630 residue [20]. This hypothesis suggests that compound 4 may have an expanded multitarget effect, which should be verified experimentally in future investigations.
Molecular docking studies
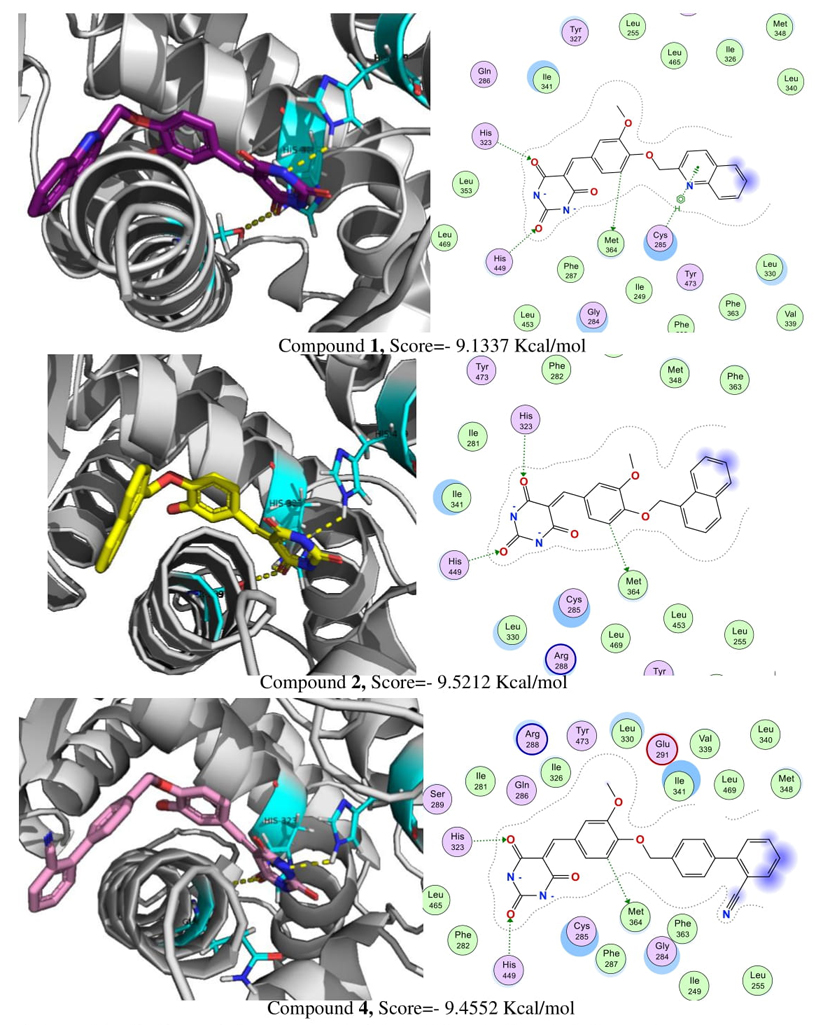
Based on in silico pharmacological consensus analysis (PHACA) compounds 1, 2 and 4 were chosen to test their in vitro expression on PPAR-γ and GPR40 and their activation products GLUT-4 and insulin, respectively. To establish a correlation between their presumed binding patterns and their experimental activities, molecular docking was conducted on these targets. To validate our molecular docking model, the Root Mean Square Deviation (RMSD) was calculated as a measure of re-docked success of rosiglitazone over PPAR-γ with a value of RMSD =1.0369 Å and a score of −8.4423 Kcal/mol. The molecular docking reveals that compounds (Fig. 4) 1, 2 and 4 occupied the ligand binding pocket of PPAR-γ and form hydrogen bonds with His-323 and His-449, essential interactions for the activation of this receptor, in addition an interaction with Met-364 was observed in all compounds. However, compound 1, exhibiting the highest activity in vitro, demonstrated an additional H-arene interaction with Cys-285. This interaction has been linked to conformational changes in PPAR-γ that could potentially contribute to its activation [21].
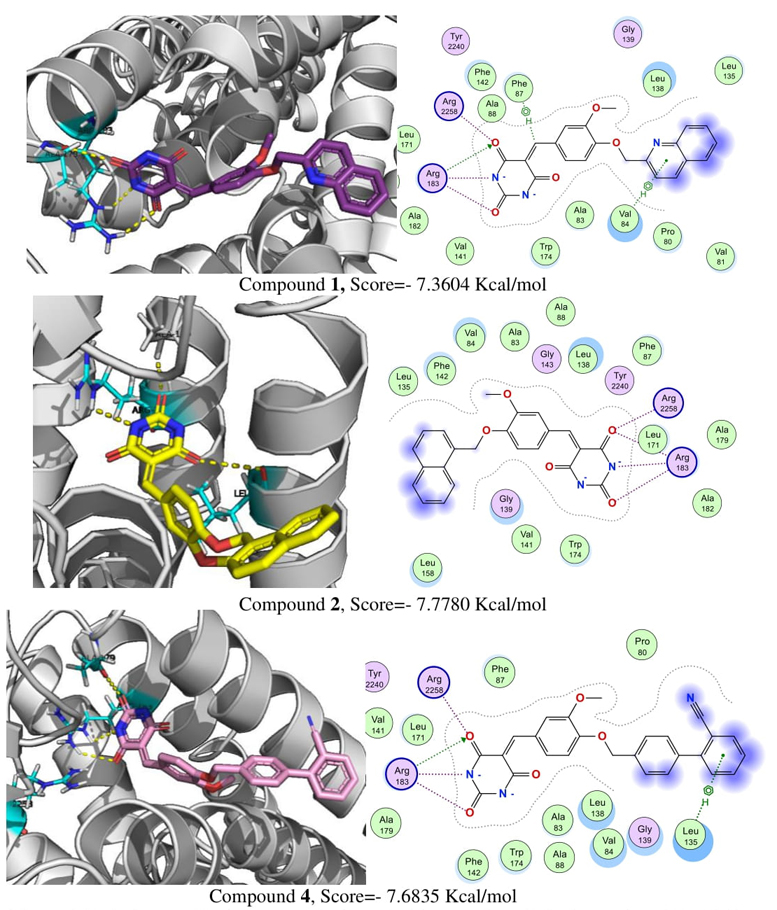
With respect of GPR40, the validation with co-crystal molecule TAK-875 has a RMSD =0.4234 Å and a score of −11.3756 Kcal/mol.
Molecular docking on GPR40 (Fig. 5) suggests that compounds 1, 2 and 4 established electrostatic and hydrogen bonds with Arg-183 and Arg-258 residues, which are characteristic of GPR40 allosteric agonists [22]. Additionally, compounds 1 and 4, were the most potent in the in vitro and in vivo assays, showed an additional H-arene interaction with Phe-87, Val-84 and Leu-135. This could improve the interaction and affinity of the compounds with the GPR40 binding site and explain their experimental biological activity.
Conclusions
In conclusion, taken together these results suggest that benzylidenebarbiturates (compounds 1-5) showed better predicted pharmaceutical properties than the hydantoin isosteres (compounds 6-10), behaving as safe computational hits through a pharmacological consensus analysis. Compounds 1, 2, and 4 have been recognized as experimental multitarget modulators of PPAR-γ, GLUT4, and GPR40 proteins, displaying in vivo antihyperglycemic effects alongside predicted pharmacokinetic and toxicological profiles conducive to their potential as antidiabetic candidates.











 nueva página del texto (beta)
nueva página del texto (beta)