Introduction
Gastrointestinal diseases represent one of the main public health problems that affect people of any age and social status; however, the most vulnerable groups are children and the elderly. Diarrheal diseases are the second cause of disease in children under 5 years.
In 2017, the World Health Organization1 estimated that 1.7 million episodes of diarrhea occur each year in developing countries. Diarrheal diseases kill 525,000 children under the age of 5 each year. Worldwide gastrointestinal infections are one of the most important causes of morbidity and mortality among infants and children2.
A large group of viruses, parasites, and bacteria cause diarrheal diseases. Within the latter, the most common bacterial etiological agents are considered Escherichia coli, Vibrio cholerae, Salmonella sp., Shigella sp., Campylobacter fetus, Campylobacter jejuni, and Yersinia enterocolitica.
E. coli colonizes the intestine of humans a few hours after birth and is considered part of the normal microbiota, but there are pathogenic strains that can cause intestinal and/or extraintestinal damage, which leads to different syndromes one of them being diarrheal syndrome3.
The diarrheagenic E. coli is the major cause of diarrheal diseases in both developed and developing countries. In developing countries, these are responsible for the high morbidity and mortality in children under 5 years of age4,5. According to reports from the World Health Organization (WHO), acute diarrhea caused by E. coli finds itself in second place next only to rotavirus6,7.
In Mexico, according to the data of the morbidity yearbook of the General Directorate of Epidemiology of 2013, diarrheal diseases are in second place among the 20 leading causes of illness in children under 5 years of age.
The pathogenic strains of E. coli have virulence factors that can be classified as adhesins and toxins. The adhesins allow bacteria to colonize the intestine; they can be divided into two groups: fimbrial adhesins and afimbrial adhesins. Afimbrial adhesins are outer membrane proteins such as the case of intimin4,8. In relation to toxins, they are classified into two groups, endotoxins (lipopolysaccharide, LPS) and exotoxins. Exotoxins are classified as enterotoxins, cytotoxins, and neurotoxins. Enterotoxins produce watery diarrhea due to the release of water and electrolytes by the enterocyte, and the cytotoxins produce inflammatory diarrhea whose stools contain mucus and blood3.
The fimbrial adhesins allow the union of the bacteria to the recipients of the microvilli of the enterocytes; this is followed by the secretion of toxins that are responsible for the production of diarrhea. Once E. coli has colonized the intestine, diarrhea occurs, it can be watery or inflammatory, watery diarrhea is due to the production of one or more enterotoxins and inflammatory diarrhea to the production of cytotoxins3.
E. coli strains based on their mechanism of damage are classified into six pathotypes: enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enterohemorrhagic E. coli (EHEC), diffuse adhesion E. coli (DAEC), and enteroaggregative E. coli (EAEC)4,8.
The ETEC strains represent the main cause of diarrhea in children and are the cause of the so-called traveler's diarrhea. The virulence factors of ETEC are bacterial adhesins and enterotoxins. This group is characterized by the presence of one or more plasmids coding for thermolabile enterotoxins (encoded by the lt gene) and thermostable (encoded by the st. gene) which include type I (STa) and type II (STb)3.
This pathotype is characterized by a pattern of localized adherence (AL) to HEp-2 cells, forming microcolonies. The adherence of EPEC is associated with the presence of the 50-70 MDa E. coli adhesion factor (EAF) plasmid (in English, EPEC adherence factor). The adherence of the EPEC in the intestine is called adherence and effacing (A/E) and is caused by the intimate union of the bacteria to the membrane of the enterocytes; this gives rise to the polymerization of actin with the subsequent formation of a pedestal. Once the microcolonies are formed, the second stage of adherence begins and is characterized by the destruction of the intestinal microvilli, which is called sloughing. The lack of microvilli, the polymerization of the actin, and other factors induce the exit of water and electrolytes to the luminal space9.
The main virulence factors found in the EPEC strains are encoded in an island of chromosomal pathogenicity of 35 kb called locus of enterocyte effacement (LEE). LEE is organized into five large polycistronic operons called LEE1 to LEE5 where the products are: a type III secretion system (LEE1 to LEE3), a protein translocation system (LEE4), and an adhesion system consisting of a protein membrane of 94 KDa denominated intimina (EAE of the English E. coli attaching and effacing protein), which is responsible for the intimate adhesion between the bacterium and the epithelial cell, as well as a translocated intimin receptor receiver) encoded by LEE51110,11.
The adhesion also involves a type IV pili or curled fimbrias called bfp (from English, bundle-forming pilus), whose genetic information is encoded in the plasmid "EAF". This also codes for a transcriptional activator called "regulator encoded in the plasmid" (Per) that is required for the activation and optimal function of genes encoded in LEE and for the expression of Bfp12. The bfpA gene is responsible for the AL phenotype, characterized by the ability of bacteria to form microcolonies on the surface of intestinal epithelial cells13,14.
The main characteristic of EAEC strains is the ability to produce a pattern of aggregative adherence (AA) in HEp-2 cell culture, similar to "brick stacking"4. Strains of this phenotype possess the ability to bind both HEp-2 cells, as well as the surface of glass lamellae. Among the virulence factors are the aaf fbrial genes, which are encoded in a plasmid pAA; the aatA gene that corresponds to a fragment of the CVD432 gene which codes for an outer membrane protein (Tol C), component of an ABC transporter that participates in the secretion of the dispersion encoded by the aap gene15-18. Some EAEC strains are capable of producing the enteroaggregative thermostable toxin encoded by the astA gene19. These strains produce lesions known as adhesion and A/E sloughing due to the presence of the eaeA gene that codes for intimina15.
The EHEC pathotype includes strains of different serotypes that present the same pathogenic characteristics of serotype H7:O157, which is considered as the prototype of the group. This is very close to Shigella sp and invades the epithelium of the colon. Once inside the intestinal cells, bacteria synthesize toxins. This pathotype has been associated with the etiopathogenesis of afebrile hemorrhagic colitis (CH), hemolytic uremic syndrome, and thrombocytopenic purpura. The strains that cause these syndromes produce one or more cytotoxins currently called Shiga toxins (Stx1 and Stx2). Other damage caused by this group of bacteria is known as adherence and effacing (A/E). The main mechanism of pathogenicity of EHEC is the production of cytotoxins (Stx) encoded in the bacteriophage STX inserted in the bacterial genome, and the bacteria can synthesize one or both toxins8.
The EIEC and Shigella sp. pathotype are related genetically and biochemically3. However, diarrhea produced by EIEC is less severe than that produced by Shigella sp. The infections produced by EIEC occur in the colon mucosa and the symptoms begin with watery diarrhea and progress to inflammatory diarrhea with blood and mucus. One of the genes related to the mechanism of invasiveness is the so-called ial that is found in the pInv plasmid, where certain proteins related to the invasion process are also encoded such as the Ipa proteins and their transcription regulator invE4. DAEC group is one of the most recently classified bacterial groups and affects enterocytes of the small intestine and the damage manifests itself as a cellular projection similar to a finger that covers the bacterium15. The mechanism of pathogenicity is clearly unknown. However, some virulence factors have been described, such as the Afa/Dr adhesins. These strains show a diffuse adherence pattern since they do not form colonies when they adhere to Hep-2 cells20. The strains of E. coli belonging to the DAEC pathotype have been epidemiologically linked to diarrheal syndromes, particularly in children under 12 months of age15.
Recently, in several countries of Latin America and North America, epidemiological studies have been carried out whose purpose was to determine the different pathotypes associated with cases of childhood diarrhea. These studies have shown that strains of E. coli that belong to the ETEC and EPEC types are the main isolated pathogens8,21-23. In the case of Mexico, data published from the year 1987 to 2009 demonstrate the presence of EPEC in most of the isolates obtained from infected patients. A prevalence of 17-19% infant diarrhea cases in different regions of the country was noted24. Recently, Estrada García et al. reported the presence of four different pathotypes of E. coli in a peripheral community in Mexico City. In a cohort study in children under 2 years, interestingly in 41% and 38% of the 32 samples obtained with episodes of acute diarrhea, associations were found with atypical EPEC and ETEC, respectively. It is also notable that the majority of diarrheagenic infections were associated with asymptomatic events.
Antimicrobial resistance represents a public health problem, identified by the WHO25 as one of the main health problems. A new report from the WHO reveals that this serious threat is no longer a forecast for the future and is already in all regions of the world a reality that can affect any person of any age in any country.
Among the pathogens emerging and resistant to antimicrobials are E. coli, which affects humans and different animal species. Research in both humans and animals shows resistance to the most frequently used antimicrobials26, in addition to which, bacteria are generally resistant to more than one antimicrobial27. Studies in Mexican children show that the bacteria that cause diarrheal disease in hospitalized children are resistant to trimethoprim-sulfamethoxazole (TMP-SMX) and ampicillin, which are the drugs most commonly used in the treatment of pediatric diarrhea21.
Materials and methods
Bacterial strains and characterization
A total of 284 strains of E. coli obtained from 151 stool samples from children with diarrhea, < 5 years old, were isolated. Of the 151 samples, 81 came from children's samples and 70 from girls' samples.
The stool samples were obtained by highly trained medical personnel of the National Institute of Pediatrics, which were later transported to the laboratory in the Cary Blair® transport medium, where they were planted in MacConkey Agar (DIBICO®) and incubated for 24 h at 37°C. After this time, one to five colonies were selected that fulfilled the morphological characteristics of E. coli, which were subjected to conventional biotyping tests to identify the bacterial species. To evaluate b-hemolysis, these strains were seeded on blood agar and incubated at 37°C for 24 h.
Reference strains
A panel of E. coli reference strains was used as positive controls for the different polymerase chain reaction (PCR) assays, antibiograms, and adhesion to HEp-2 cells. Strains TB334C (Stx1+, Stx2+, eaeA+), B-171 (BfpA+, eaeA+), EIEC O124:H30 (ial+), and O407:H1 (LT+, ST+) were kindly donated by Dr. Estrada García of CINVESTAV. EAEC 044:H18 (aat and astA), strain donated by Dr. Chattaway of Public Health England. Finally, the National Institute of Epidemiological Reference of Mexico (INDRE) provided strains O42 and C600.
DNA extraction and PCR reaction
The genomic DNA of the E. coli isolates was extracted by the boiling method, previously described by Al-Gallas et al., 200728. The PCR (single or multiple) allowed to detect genes that code for toxins (LT, STa, Stx1, Stx2, and EAST1) and genes that code for adhesins (intimina, eae, pili type IV, bfpA, for invasiveness and gene encoding for dispersin, and aat) (Table 1). The PCR reactions used for the amplification of the different genes were carried out using Apex Taq DNA polymerase (Genesee Scientific, USA) and primers. The 50 µl reaction mixtures contained the following: 1X buffer (100 mM Tris-HCl, pH 8.5, 500 mM KCl, 15 mM MgCl2, 1% Triton X-100), 0.2 mM of each dNTP (Apex dNTP mixture, 10 mM), 0.25 µM of each primer, 2.0 U of Apex Taq DNA polymerase, 3 µl of tempered DNA, and 37 µl of nuclease-free water. The multiplex PCR products (mPCR) were separated on a 2% Agarose gel (Amresco, Ohio, USA). The amplified PCR products were visualized with ethidium bromide under a UV illuminator and recorded using the Digidoc Imaging System (UVP, CA, USA).
Antimicrobial susceptibility assays
The antimicrobial susceptibility test was carried out to determine the profiles of susceptibility and resistance of the bacteria to the antibiotics tested. For this, the paper disc diffusion method was used and consists of impregnated antimicrobial discs with ampicillin (10 µg), amikacin (30 µg), kanamycin (30 µg), cephalothin (30 µg), amoxicillin/clavulanate (30 µg), cefotaxime (30 µg), TMP/SMX (1.25 µg/23.75 µg), and cefoxitin (30/mg). These discs (6 µm diameter) were placed in an automatic Sensi Disc dispenser (DB BBL Sensi-Disc Antimicrobial Susceptibility Test Discs, Sparks MD, USA). The inoculated agar plates were incubated at 37°C for 18-24 h. After incubation, the diameters of the zone of inhibition of the antibiotic were measured. The results obtained were used to classify the isolates as resistant, intermediate resistant or susceptible to a particular antibiotic using standard reference values according to the indications of the National Committee of Clinical Laboratory Standards of 2014. The resistance to multiple drugs is defined as resistance to three or more different classes of antibiotics.
Adhesion test in HEp-2 cells
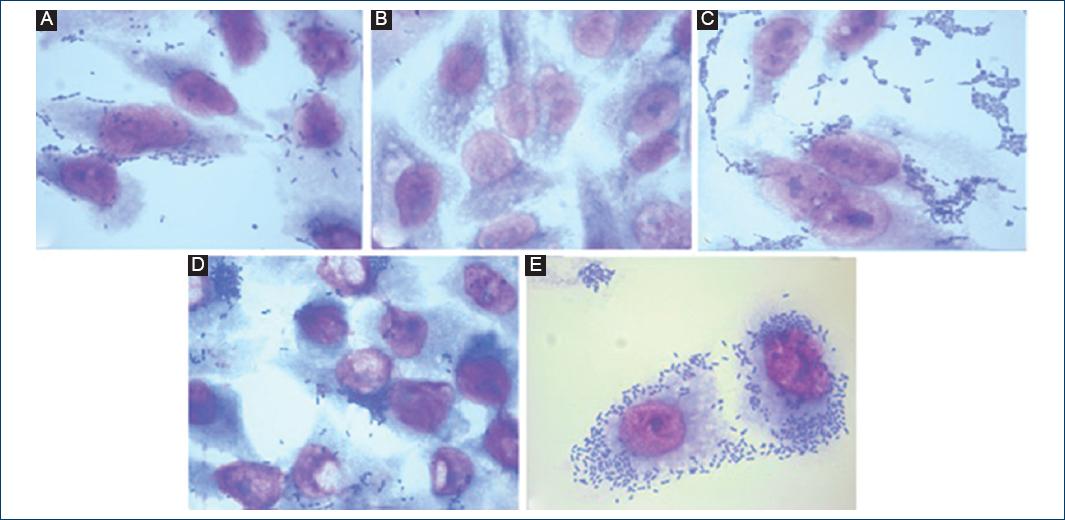
Fifty-six strains of E. coli previously identified as EAEC (ast1+ and/or aat+) were tested for adhesion to HEp-2 cells as described by Donnenberg and Nataro in 199529, with modifications. In summary, 6.0 × 105 HEp-2 cells were cultured in DMEM medium with high glucose content (Biowest, Miami FL, USA) supplemented with 10% fetal bovine serum (Biowest, Miami FL, USA) and 1% antibiotic/antifungal solution (PAA, Pasching, Austria). The cells were placed in 24-well plates (TPP, Switzerland) containing a glass lentil and incubated until reaching a confluence of 80-90% (1-2 days). The HEp-2 cells were washed 3 times with sterile phosphate-buffered saline (PBS), and 500 µl of fresh DMEM medium was added without fetal bovine serum or antibiotics.
One day before the experiment, the E. coli strains were grown overnight in 1.5 ml of LB medium with 1% D-mannose and the concentration was adjusted to 0.5 UMF (2 × 108 CFU/ml) with sterile PBS. The cultures were centrifuged and then resuspended in 1 ml of sterile PBS. 50 µl of the bacterial suspension was mixed with 450 µl of DMEM medium without fetal bovine serum or antibiotics and inoculated into cultures of HEp-2 cells. The infected cells were incubated for 3 h at 37°C under a 5% CO2 atmosphere and washed 5 times with sterile PBS, fixed with absolute methanol for 1 min and stained with 10% Giemsa. Adherence patterns were assigned according to the description of Nataro and Kaper in 19984. E. coli O:42 strains showing aggregate adhesion and E. coli K:12 not binding to HEp-2 cells were included as controls in each experiment.
Results
A total of 284 strains of E. coli obtained from 151 stool samples from children with diarrhea, under the age of five were isolated. Of the 151 samples, 81 came from samples from children and 70 from samples from girls (Table 1)
Of the 284 strains of E. coli isolated from feces of children with diarrhea, only 179 were positive for at least one of the nine genes studied. Of these, there was a higher prevalence of genes coding for LT toxins (78/179, 43%), ST (50/179, 28%) followed by astA (43/179.24%,) STx1 (36/179.20%), and Stx2 (22/179.12%), respectively. While the genes encoding both adhesins as well as other virulence factors, the ial gene (38/179, 21%) was found and codes for invasiveness had a high prevalence followed by the gene that codes for the intimina eaeA (32/179.18%) and the bfpA gene (12/179.7%) that codes for molecules related to the transport of the protein bundlina. The gene with the lowest prevalence was aat (6/179.3%), which codes for the anti-aggregative protein carrier formerly called CVD432, which has been associated with tEAEC strains (Fig. 1).
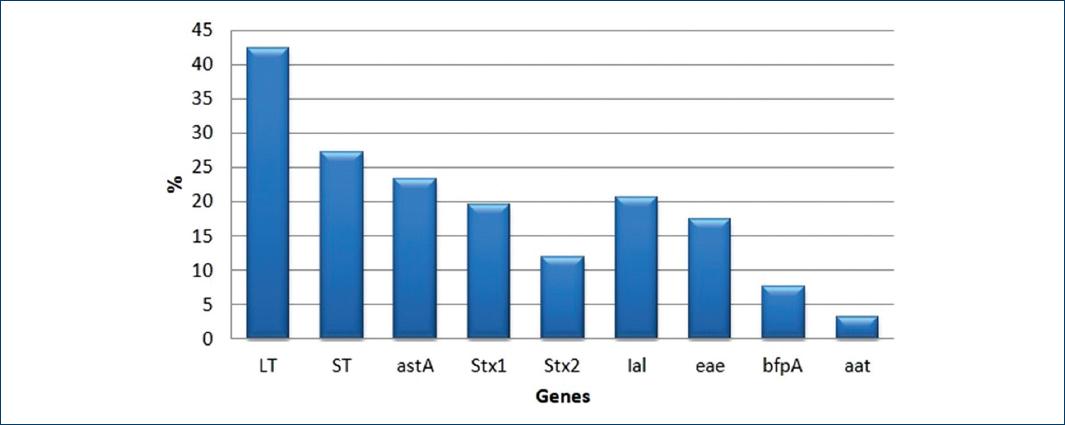
Figure 1 Percentage of Escherichia coli genes, found in the 179 positive strains for at least one gene.
Identification of E. coli pathotypes
The E. coli pathotypes were identified based on the Estrada-García et al., 2009, classification. One hundred and thirteen belong to ETEC, 45 to EHEC, 45 to EAEC, 38 to EIEC, 19 to aEPEC, and 2 to tEPEC. This number virotypes are due to an E. coli strain sharing genes with another strain of E. coli (Table 2 and Fig. 2).
Table 2 Virulence genes and E. coli virotypes
| Virotype | *Number of positive strains | % (n = 179) | Identified genes (number of positive samples) |
|---|---|---|---|
| ETEC | 113 | 63 | LT (63) ST (36) LT:ST (14) |
| EHEC | 45 | 25 | Stx1 (17) Stx2 (8) Stx1:eaeA (6) Stx1:Stx2 (10) Stx2:eaeA (2) Stx1:Stx2:eaeA (2) |
| EAEC | 45 | 25 | aatA (2) astA (39) aatA:astA (4) |
| EIEC | 38 | 21 | ial (38) |
| aEPEC tEPEC | 19 2 | 11 1 | eaeA (19) eaeA:bfpA (2) |

Figure 2 Escherichia coli pathotypes isolated from children with diarrhea who share different genes.
Of the 179 strains of E. coli, 115 of them (64%) were positive only to one virotype, 45 (25%), 17 (10%) were positive to two and three virotypes, respectively. Only 2 samples were positive to 4 virotypes (Table 3).
b-hemolysis
Of the 179 strains obtained, only 37 of them showed complete hemolysis, and only 16 had at least one virulence gene.
Distribution of virotypes by sex
The EHEC and ETEC virotypes were presented in a greater percentage in the samples from children with 63% and 57%, respectively, in relation to those obtained from girls (37% and 43%). However, the EAEC (56%) and aEPEC (58%) virotypes were observed more frequently in girls. The tEPEC virotype was only observed in two samples, one from each group.
The EIEC virotype was observed in 50% of the samples from both girls and boys.
Identification of E. coli virotypes according to the seasonal stage
In relation to the seasonal period, in the spring and summer, 45.8% and 30.6% of the samples were collected, respectively. In autumn and winter, only 23.6% of the samples were collected.
Of the total E. coli strains isolated from 113 ETEC virotypes, 52% were obtained in summer, 25% in spring, 14% in autumn, and 8% in winter. This incidence was also presented for EAEC. The EHEC virotype was the only one that did not show a trend since all the samples were collected throughout the four seasons of the year. As for the 38 EIEC virotypes, the behavior was different since 37% of the strains were collected in winter followed by summer with 29%, autumn with 24%, and finally spring with 10%. In relation to the EPEC virotype, a greater presence of strains can be observed in the summer season, followed by winter with 25%, autumn 17%, and spring 5%.
Adhesion tests to HEp-2 cells
For this trial, only the strains that presented the aat and/or astA genes for the EAEC virotype were included and eaeA and/or bfpA for the EPEC virotype. Typical EPEC strains (tEPEC) were considered those that were found to be eae:bfpA positive and atypical EPEC (aEPEC) strains that were positive only for the eaeA gene.
Of the 45 strains obtained with the EAEC virotype, the strains astA + and/or aat+ either had mainly aggregative and diffuse adhesion (33/45, 73%). Only three strains that were astA positive showed localized adherence.
Of the 21 strains that presented the EPEC virotype, 19 were aEPEC, of which 12 of them (63%) had some type of adherence with diffuse adherence being the most observed. About 37% of the strains did not present any type of adherence to HEp-2 cells. Of the two strains of tEPEC, only one presented localized adherence and other diffuse adherence (Table 4 and Fig. 3).
Table 4 Adherence of EAEC and EPEC virotypes to HEp-2 cells
| EAEC | Total samples | *AA (%) | *DA (%) | *LA (%) | *NA (%) |
|---|---|---|---|---|---|
| astA | 39 | 12 (31) | 16 (41) | 3 (8) | 8 (20) |
| aat | 2 | 1 (50) | 1 (50) | 0 | 0 |
| astA:aat | 4 | 3 (75) | 0 | 0 | 1 (25) |
| aEPEC eaeA | 19 | 2 (10) | 7 (37) | 3 (16) | 7 (37) |
| tEPEC eaeA:bfpA | 2 | 0 | 1 (50) | 1 (50) | 0 |
*AA: aggregative adherence; *DA: diffuse adherence; *LA: localized adherence; *NA: non-adherent; EAEC: enteroaggregative E. coli; EPEC: enteropathogenic E. coli; E. coli: Escherichia coli.
Antibiogram
The antibiogram test was performed on the 284 strains of E. coli to determine the sensitivity to the different antimicrobials evaluated. As can be seen in Table 5, the results showed that there is a broad resistance of E. coli to ampicillin, SMX-TMP, amoxicillin-clavulanic acid, and mainly to cephalothin (Fig. 4).
Table 5 Results of antibiogram in 284 diarrheagenic E. coli strains
| Number of strains | |||
|---|---|---|---|
| R* (%) | I** (%) | S*** (%) | |
| K | 30 (11) | 55 (19) | 82 (29) |
| AN | 11 (4) | 32 (11) | 242 (85) |
| AM | 195 (69) | 36 (13) | 53 (19) |
| CF | 85 (30) | 65 (23) | 19 (7) |
| AMC | 117 (41) | 73 (26) | 77 (27) |
| CTX | 67 (24) | 23 (8) | 187 (66) |
| STX | 126 (44) | 4 (2) | 153 (54) |
| FOX | 16 (6) | 15 (5) | 228 (80) |
| CFZ | 109 (38) | 11 (4) | 94 (33) |
| FEP | 85 (30) | (0) | 124 (44) |
| CRO | 96 (33) | 3 (1) | 110 (39) |
| CIP | 91 (32) | 5 (2) | 113 (40) |
*Resistant,
**Intermediate,
***Sensitive; AM: ampicillin; AMC: amoxicillin/clavulanic acid; AN: amikacin; CF: cephalothin; CFZ: cefazolin; CIP: ciprofloxacin; CRO: ceftriaxone; CTX: cefotaxime; FEP: cefepime; FOX: cefoxitin; K: kanamycin; STX: sulfamethoxazole/trimethoprim.
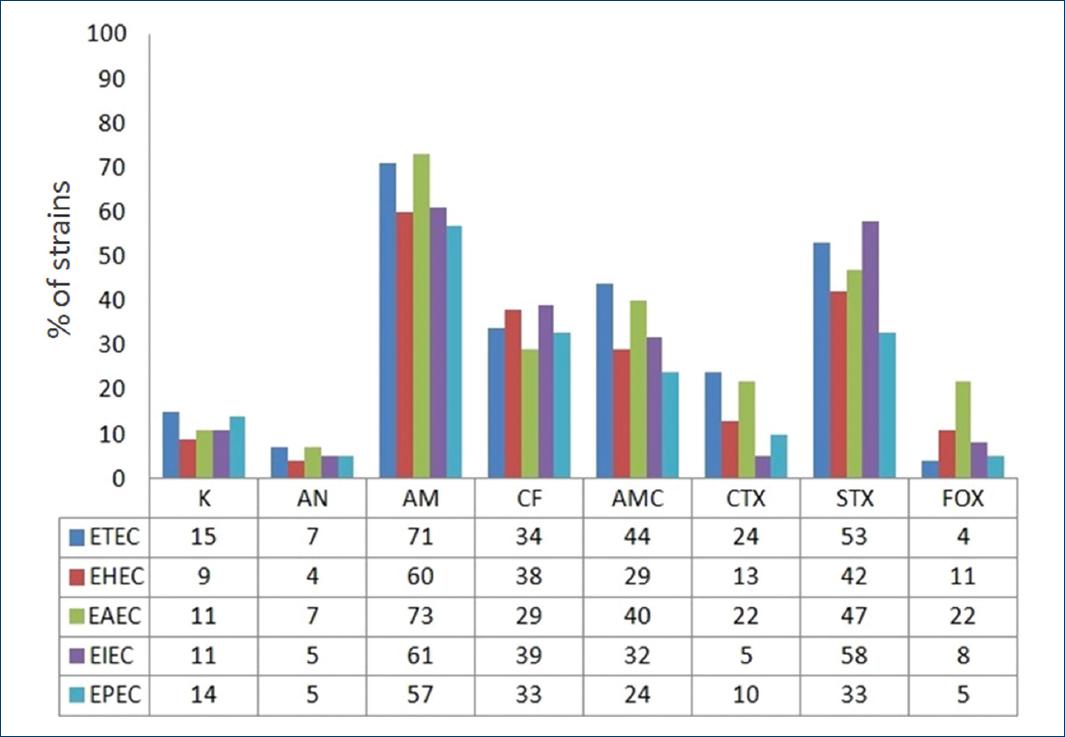
Figure 4 Antibiotic resistance pattern of the different diarrheagenic Escherichia coli virotypes. AM: ampicillin; AMC: amoxicillin/clavulanic acid; AN: amikacin; CF: cefalotin; CTX: cefotaxime; FOX: cefoxitin; K: kanamycin; STX: sulfamethoxazole/trimethoprim.
The resistance to ampicillin was 73% in EAEC strains, 71% in ETEC, 61% in EIEC, 60% in EHEC, and EPEC in 57%. Regarding SMX/TMP EIEC showed resistance in 58%, ETEC in 53%, EAEC in 47%, EHEC in 42%, and EPEC in 33%. Amoxicillin-clavulanic acid ETEC showed resistance in 44%, EAEC in 40%, EHEC in 29%, EIEC in 32%, and the EPEC virotype in 24%. Different virotypes also showed resistance to cephalothin between 39 and 29%.
Discussion
A total of 284 strains of E. coli, isolated from 151 stool samples from children with diarrhea younger than 5 years, were analyzed. Of the 151 samples, 81 (54%) are from boys and 70 (46%) from girls. These data coincide with the studies done in Nicaragua, where 60.8% of the samples were from boys and 39.2% from girls30, as reported by Cortéz in 200230 and Estrada García in 200523. They revealed that there is an association between intestinal infections and sex, independently that in this work, the difference between boys and girls with diarrhea is small, apparently, boys were more affected by enterobacteria. In relation to age, enterobacterial infection preferentially affects boys or children under 2 years of age, data that agree with those published by Kaper et al. in 200415. This is probably due to hygiene conditions since their immune system is still. It is not fully developed.
In relation to the presence of virulence genes in the E. coli isolates, of the 179 strains of E. coli that were positive for at least one of the nine genes studied, 113 strains presented the ETEC virotype. These data differ from that published by different Mexican authors (Vidal, 2007; Estrada García, 2009) who found a higher prevalence of the EPEC pathotype. Of this ETEC pathotype, 63 (55.7%) of the strains carried the gene that codes for the LT toxin, 36 (19.6%) for ST, and 14 (7.6%) for LT-ST. These results agree with that reported by Cortés-Ortiz in 200230 in Mexico and Chandra et al. 201231 in India. These results are not concordant with those reported in Bolivia, where LT (40.5%) and ST (39.5%) were detected with the same frequency, while strains carrying genes for LT-ST were detected with a lower frequency32. The virotypes that carry the gene that codes for LT are frequently found in populations of Latin America in comparison to Asia and Africa where the predominant gene is the one that codes for ST33,34.
Due to the clinical importance of ETEC, this pathotype is considered as a bacterial group of special attention by the WHO as in association with rotavirus it causes 80% of the cases of acute diarrheal diseases worldwide.
After ETEC, the pathotypes most frequently found in this work were STEC and EAEC; with 45 strains, which is equivalent to 25%, respectively, of the total of strains of E. coli isolated. Of STEC, 11 carried the eaeA gene that codes for intimin, related to the A/E lesion, which in theory would increase the virulence of this virotype. The results obtained differ from the studies carried out by Estrada-García et al., 200921 and Cortez et al., 200230. In their work, STEC was presented in percentages lower than 4%. It is considered that the incidence of this pathotype in developing countries is low, since this particular strain only appeared in industrialized countries. For example, in the USA there are 20,000 infections caused by STEC in specific by O157:H7 strain annually35. However, recently, it has been described that this disease occurs in both developed and developing countries. The studies carried out in Chile found that the STEC strains in 201214 presented a greater proportion than the STEC strains isolated in 2011. In this case, the authors consider that such an increase in the number of isolates of STEC strains implies an emerging pathogen in developing countries, which may be due to the increased communication and commercialization worldwide since the said pathogen had been identified in water, sausages, and handmade yogurts, among others. This would increase infections by this pathotype, especially in countries like ours.
In relation to the EAEC virotype, with a percentage of 25%, there are studies that show that this virotype is responsible for the majority of diarrheal infections in developing countries and stands as second cause of traveler's diarrhea31,36,37 in Peru, where EAEC was isolated in 12.9% of the total analyzed samples.
Of the isolates of E. coli, the EIEC pathotype was present in 20.7% of the total strains. A fact that differs with that reported in Mexico as well as in other developing countries such as Vietnam38. In Vietnam, this pathotype was identified in 2% of the isolated strains and in Tanzania, this pathotype was not detected39. Regardless that only lactose-positive colonies were isolated in this study, the percentage of this pathotype is very different from that reported in studies conducted in Mexico and other countries, without taking into account that 60% of E. coli strains belonging to the EIEC pathotype are lactose negative4.
Interestingly, the E. coli pathotype that was presented in low percentage was EPEC with 11.37% of the studied strains unlike that published by several Mexican authors and Rajendran, in a study conducted in India in 201040, who found EPEC in 10% of strains. Atypical EPEC was presented in 90% (i.e., 19 of the 21 strains) and typical EPEC in only two cases, which is equivalent to 10%, a result that coincides with that published by Estrada, 200521 and differs with that published by Afset et al. in 201141 and Nguyen et al. 200642. They mention that atypical EPEC is more frequent in industrialized countries. On the other hand, Estrada-Garcia in 200521 suggests that atypical EPEC is an emerging pathogen not only in industrialized countries but also in developing countries.
In relation to the seasonal period, the period in which more strains of E. coli were isolated was in summer 69 (45.8%), followed by 46 (30.6%) in spring, 20 (13%) in winter, and 16 (10.6%) in autumn. The presence of E. coli in the samples obtained was in the hot and humid months. The ETEC pathotype was found mainly in the warmer months, from June to September, which coincides with what was published by Estrada-García in 200921, who found a peak in the ETEC isolates from June to October. Estrada-García suggests that humidity is an important factor for the propagation of ETEC in Mexico City. As for EPEC, this strain is presented in summer and winter. In 2009, Estrada-García found that EPEC is distributed throughout the year, mainly from April to October, with a peak in June. A study conducted in Norway found that EPEC was isolated from children under 5 years of age throughout the year but was found most frequently in late summer and early fall43. This variation in data may be due to different factors such as place of origin of children, eating habits, overcrowding, and characteristics of the place such as temperature and humidity, among others. In our study, the samples come from a busy hospital setting attended by children from Mexico City as well as from other states of Mexico.
In relation to the virotypes identified in E. coli strains, the presence of a high percentage of genes shared by these strains is striking. This genetic heterogeneity suggests the existence of a high horizontal gene transfer potential, which was already described by Donnenberg, 200243 in strains of E. coli from samples of children with diarrhea of genes shared by different pathotypes.
Regarding the resistance to different antimicrobial agents, the 284 strains isolated from E. coli showed a higher percentage of resistance to four of the eight antibiotics tested. These were: ampicillin, cephalothin, amoxicillin-clavulanic acid, and SMX-TMP. A fact that coincides with that published by Literak et al. in 201144 who found great resistance mainly to ampicillin. In 2005, Estrada-Garcia21 reported that ampicillin and SMX-TMP are antibiotics that have greater resistance in E. coli strains in Mexico. These results may be attributable to the indiscriminate use of antibiotics.
In relation to the pathotypes, ETEC presented a multiresistance to ampicillin, cepflotin, and mainly to TMP-SMX. This is probably related to the use of these antibiotics in Mexico during the recent years21. The results of the antibiogram performed on the STEC pathotype resulted in multiresistance to cephalothin, ampicillin, and TMP-SMX, respectively. These data agree with the results presented by Fazeli and Salehi in 200745, who showed that strains of EHEC isolated from children with diarrhea had a high resistance to penicillin derivatives as well as to TMP-SMX. In relation to EAEC, this pathotype showed greater resistance to ampicillin, followed by cephalothin and amoxicillin/clavulanic acid. This data agree with that described by Sobieszczańska et al. in 200346, who reported that EAEC can have a moderate to high level resistance to ampicillin because these drugs are recommended by the WHO for treatment in persistent diarrhea47,48. EPEC in general presented a lower percentage of multiresistance, which suggests that EPEC strains could be acquired "recently" in this country. The resistance of E. coli strains does not occur for a single antimicrobial agent as can be observed in the present work. In most strains a multi-resistance were presented, which makes treatment even more difficult.
Despite the efforts made for the prevention and control of infectious diseases, they continue to represent a global public health problem. Changes in society, technology, and microorganisms by themselves contribute to the emergence of new diseases, re-emergence of already controlled diseases and the development of antimicrobial resistance. Effective control of infectious diseases in the new millennium will require effective public health strategies that have infrastructure to respond and prevent emerging health problems. The emergence and spread of antimicrobial resistance have been documented as a serious global problem.











 nueva página del texto (beta)
nueva página del texto (beta)