Introduction
The influenza A virus (IAV) belongs to the Orthomyxoviridae family and is classified into subtypes based on two surface glycoproteins, haemagglutinin (HA) and neuraminidase (NA). To date, 16 HA subtypes and 9 NA subtypes have been detected in wild birds.1 Migratory wild ducks are considered the main natural reservoir of IAV. The prevalence of IAV in ducks in North America varies from less than 1% during the spring migration to 30% prior to and during the fall migration, but large variations in virus prevalence have been observed in different surveillance studies.2-4 The virus has been isolated from 20 of the 42 species of ducks, geese and swans native to North America.5 The highest prevalence of IAV has mainly been reported in dabbling ducks of the Anatidae family and the Anatini tribe, and the majority of IAV isolates have been reported in mallard ducks (Anas platyrhynchos).6 The highest prevalence of IAV occurs in young ducks during the period prior to migration, which corresponds to late summer and early fall, while the lowest prevalence occurs during the autumn-winter period in both regions of Eurasia and North America.7 IAV transmission among wild duck populations occurs through the faecal-oral route by ingesting water contaminated with the virus. Once the virus is in the intestinal tract, it replicates and is then expelled into the environment along with faeces.8
Since the appearance of the H5N1 virus in wild migratory birds in Southeast Asia, the surveillance of wild ducks has focused on regions where migratory flyways are connected or where large duck populations are present.9,10 The environment plays an important role in the circulation and persistence of IAV. Many environmental (season, temperature, species richness and abundance) and anthropic (existence of a human population, livestock and/or poultry, existence of densities and interactions, human use of wetlands, etc.) parameters can support IAV transmission,11 especially in those scenarios in which there are interactions between waterfowl, poultry, livestock and/or humans, as well as with the wider circulation of IAV in domestic animals.12,13
Most studies of IAV in wild waterfowl (duck, goose and swan) that inhabit wetlands report the number of birds sampled and the species to which they belong, leading to the idea that any wild waterfowl presents a risk. However, one species of the 173 waterfowl species14 is frequently reported in databases and literature around the world, the mallard (A. platycrhynchos).15 This is because most studies have scarce or limited descriptions of the biological features and the diversity of the duck population, which can be influenced by the purpose and size of the wetlands where viruses have been detected or isolated. Because wild ducks require an aquatic ecosystem that provides them with food and shelter as part of their life cycle, the characteristics of the natural or artificial wetland will determine the number and species of wild ducks that can be harboured, potentially influencing the quantity and diversity of the influenza A virus present in the wetland.
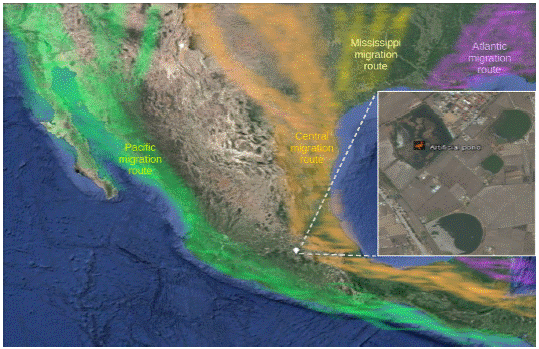
The North American Waterfowl Management Plan (http://nawmp.wetlandnetwork.ca) indicates that twenty-eight wetlands for migratory duck conservation in Mexico are visited by 27 species of migratory ducks during fall and winter through the Pacific and Central migratory routes, representing between seven and up to 17 % of an estimated population of 40 million ducks. In the central highlands of Mexico, there are smaller natural and artificial wetlands, which are isolated from each other. These wetlands provide food and shelter for migratory ducks, and they are visited by fewer than a third of migratory ducks compared to the Pacific coast and the Gulf of Mexico regions, which are visited by three-quarters of the total population. The highland region in the State of Mexico (2600 m above level sea) possesses a few hectares of artificial ponds that are used for livestock, fishing and irrigation of agricultural fields in the area; these ponds harbour migratory wild ducks during the winter because they provide temporary food and shelter. The Mexican duck (A. platyrhynchos diazi) is the only dabbling duck resident species living in these environments; the migratory dabbling ducks are Mareca strepera, Mareca americana, Spatula discors, Spatula cyanoptera, Spatula clypeata, Anas acuta and Anas crecca.16
Some poultry farms in the high plateau of Mexico have been endemic for avian influenza H5N2 low-pathogenic and H7N3 high-pathogenic strains since 2004 and 2012, respectively, while the influenza virus in wild birds has been scarcely reported by the World Organization of Animal Health. The aim of this study was to investigate whether a temporary artificial pond of 16 hectares used by humans provides an environment suitable for supporting IAV. Every year, the pond is visited by migratory ducks and is used for agriculture, fishing or livestock activities.
Materials and methods
Location
We evaluated an artificial pond of 16 hectares located in the county of Toluca in the State of Mexico (Figure 1). The pond is used for temporary agricultural irrigation during the months of March and April, and hunting is not allowed. The pond receives water from rain during the summer. The end of fall (October) marks the arrival of migratory ducks of nine species such as A. crecca and S. discors.17
Sentinel ducks
Eleven one-day-old ducks belonging to the Pekin race that were free of avian influenza virus, Newcastle disease and Salmonella spp were obtained from a commercial farm. The ducklings were fed and maintained in an isolation unit with food and water ad libitum until they were one year of age. Serological and isolation tests for both viruses and bacteria (previously mentioned) were performed to ensure that the tests were negative. The sentinel birds were introduced into the study location on September 1, 2007 (before wild bird migration), and they stayed at the centre of the wetland until March 2008; thus, the animals acted as sentinels before, during and after the stay of the migratory ducks (7 months).
Wild duck observation
To confirm the presence of wild waterfowl during the stay of sentinel ducks, we observed birds once a month through binoculars and photographs and maintained records by a simple observational-qualitative manner during the time of study.18,19
Design
The sentinel ducks were transported and maintained at the study site starting before the arrival of the migratory ducks until after their departure (September 2007 - March 2008). Samples were collected every 21 days to avoid disturbing the migratory ducks, and the samples were stored at 4 °C for transportation. Swab samples were used for chicken embryo virus isolation. The isolated viruses were tested by haemagglutination and identified and characterized by RT-PCR.
Sampling
Trachea and cloaca swab samples were separately collected for each group of ducks. The swab samples were divided into four groups. Three groups each had three swab samples from the ducks, and the other group had two swab samples from the ducks. The swabs were placed in a sterile tube with 1 ml of UTM-RT (COPAN, USA) virus transport medium. The samples were processed by centrifugation (2000 x g / 15 minutes) and filtration (0.45 nm Millipore membranes, USA); then, the antibiotic-antimycotic (penicillin G sodium salt 10,000 IU/ml, streptomycin 10,000 µg/ml and amphotericin B 25 mg/ml) (Gibco, USA) was added before inoculation in specific pathogen free (SPF) chicken embryos.
Viral isolation
Viral isolation was performed using three passes in the SPF chicken embryos.20 The haemagglutinating virus was stored at -75 °C and sent to the laboratory of the Mexico-United States Commission for the Prevention of Foot and Mouth Disease and Other Exotic Diseases of Animals (CPA) to deduce the H5 or H7 subtypes using the A/Pekin duck/Mexico/CPA-5009/2007 registration. The genome was determined based on the first isolated virus in the SPF chicken embryos.
PCR and nucleotide sequencing
Each segment of the IAV genome was amplified by PCR using previously reported primers.21 To complete the nucleotide sequences, other initiators were designed in the laboratory using the Primer3 program version 0.4.0 (HHMI & NIH, US).22 The initiators were evaluated with the NetPrimer program (Premier Biosoft, US) based on 50 sequences that were available in GenBank. The viral RNA was extracted from the allantoic fluid, which was positive for IAV (stored at -75 °C), using the PureLink Viral RNA/DNA Mini Kit (Invitrogen, USA), according to the supplier’s specifications. Each of the eight segments of the viral genome was amplified by the QIAGEN® OneStep RT-PCR kit (QIAGEN, Germany) using the designed primers for each segment. The PCR products were purified using the QIAquick Gel Extraction kit (QIAGEN, Germany). The sequencing reaction was performed for each segment by the chain termination method using the BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, USA), and the sequences were read using a 3130 Genetic Analyzer (Applied Biosystems, USA). The obtained sequences were aligned and edited to build the consensus sequences of each segment using the MEGA program version 6.23
Phylogenetic analysis
Each of the eight segments was compared with public domain sequences (Gen-Bank) using BLAST to identify similar sequences. A phylogenetic analysis of each of the eight segments of the A/Pekin duck/Mexico/CPA-5009/2007 virus genome was performed using 100 sequences with the highest similarity for each segment. The sequence alignment was processed with the Clustal W multiple alignment tool and the genetic distances were obtained by the two-parameter Kimura method. Phylogenetic trees were constructed by using the neighbour-joining method with 1,000 replicates in the MEGA program version 6.
In silico analysis of virus proteins
The amino acid sequence analysis (where it was known that there are the following mutations associated with pathogenicity: HA, NA, PB1, PB2 and NS1) was performed using the NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and GlycoEP (http://www.imtech.res.in/raghava/glycoep/submit.html) tools for predicting glycosylation sites, the Phobius prediction (http://phobius.sbc.su.se/) and TM prediction (http://embnet.vital-it.ch/cgi-bin/TMPRED_form_parser) for predicting topology, and the DAS TM filter (http://mendel.imp.ac.at/sat/DAS/DAS.html) and CFSSP (http://cho-fas.sourceforge.net/) for predicting secondary structure. All the tools are available on the Expasy portal for bioinformatics resources.
Results and discussion
In this study, IAV was isolated and characterized as the H6N2 subtype from Pekin ducks that were used as sentinels in a temporary artificial pond for agricultural irrigation.
Wild duck observation
The migratory ducks began to arrive at the artificial pond at the end of October, but A. platyrhynchos diazi was the only resident species that was present from the beginning of the study (September), starting with < 50 birds. Among the first migratory species to arrive at the pond were A. crecca and S. discors during October. In November, S. cyanoptera, A. acuta, S. clypeata, M. americana, M. strepera, and A. fulvigula arrived. All species were present from November to January; then, there were fewer species during February and March. A. platyrhynchos diazi was the only species present from September to March. The maximum number of migratory ducks was between 7000-9000 specimens from November to January. January was the month with the highest number of ducks, while October and February had fewer than 500 ducks (Figure 2).
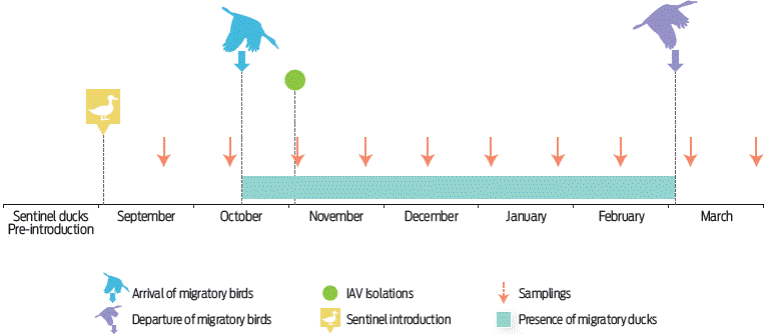
Figure 2 Schematization of the time of the study representing the sentinel ducks introduction, sampling, arrival and departure of migratory waterfowl and the detection of IAV in the pond during the study. The stay time of the migratory birds in the wetland is represented by the grey bar (based on the observational data obtained). In the figure, all important events, such as the introduction of the sentinel ducks to the pond, sampling and identification of influenza viruses, are represented in time.
The pond harboured nine species of migratory dabbling ducks and one resident dabbling species. Most IAV isolates in North America have been found in the A. platyrhynchos dabbling duck as well as in different species of geese and swans, which are not migratory species in the central highlands of Mexico.24,25 On the other hand, IAV has been reported in dabbling ducks that belong to the A. acuta, A. crecca, S. discors, M. americana, S. cyanoptera, S. clypeata, M. strepera and A. fulvigula species, which were present in this study and could be an infection source for the sentinel Pekin ducks.26-28A. fulvigula is present on the coast of the Gulf of Mexico from Florida to Tamaulipas state, which represents a limited distribution compared to the wide distribution of other migratory ducks in North America; one isolate of IAV has been previously reported for this species in Texas, USA.29,30A. fulvigula is an endangered species in Mexico and it moves to Veracruz state during the winter season.31 The presence of A. fulvigula in the artificial pond in this study is most likely accidental; therefore, its participation as an IAV reservoir is probably unlikely.
Virus isolation
One IAV subtype was isolated from the trachea and cloaca of the four Pekin duck groups on November 4th, 2007. Through serology, the CPA identified that the identified viruses belonged to the H6N2 subtype. The H6N2 virus was isolated from sentinel Pekin ducks during November, when nine species of migratory dabbling ducks were present and before the pond became populated with approximately 9000 dabbling ducks in December and prior to migration in February. The absence of the H6N2 virus or other IAVs during the months prior to or after November can be explained by the limited period of viral excretion, which lasts over seven days in migratory ducks, such as A. platyrhynchos, and by their immunity.32 The low level of detection of IAV has been observed in other conditions; for example, one H4N6 and five H3N8 isolates were found in 6/100 sentinel free-grazing ducks in rice fields in Thailand during a period of 4 months of surveillance (December 2010 - April 2011).33 Another study used domestic geese and ducks for 5 years of surveillance in the Danube delta (4125 km2), Romania. The H5N3 subtype was detected in 4 to 20/400 individuals.34 Another interesting study was done with sentinel ducks in Germany, Austria and Switzerland. This surveillance found 11 different subtypes in a period of 2 years (2006-2008) in wetlands with wild birds,35 but the authors of these studies did not distinguish between waterfowl or wild bird populations and species. A surveillance of wild mallards residing year-round in an artificial pond in Sweden found 9 of 977 faeces samples to be positive for five different IAV subtypes.36
The lack of IAV infection in sentinel ducks could also be related to the low detection of the virus in water. A study by Ito et al37 found 12 of 102 positive samples from lake water in Alaska over a period of 2 years. Another study reported the isolation of one H4N2 subtype of 240 water samples taken from a pond with wild waterfowl in Mexico.38 On the other hand, studies of IAV surveillance of lake sediment in Alaska between 2005 and 2006 recorded 45 of 91 positive samples by RT-PCR but did not perform isolation.39 A high prevalence of IAV in the water was demonstrated in ducks farmed in ponds, where 23 of 35 water samples were positive for IAV.40 Therefore, the presence of IAV found in sentinel ducks is lower in natural wetlands than that found in farmed ducks.
Sequencing of the virus genome and phylogenetic analysis
The complete sequence of the complete genome of the isolated virus in the SPF chicken embryo was performed. The nucleotide sequences from each of the eight segments of A/Pekin duck/Mexico/CPA-5009/2007 were entered into the GenBank database with the following accession numbers for each segment: basic polymerase two [KP179319], basic polymerase one (PB1) [KP179320], acid polymerase (PA) [KP179321], haemagglutinin (HA) [KP179322], nucleocapsid (NP) [KP179323], neuraminidase (NA) [KP179324], matrix protein (M) [KP179325], and nonstructural protein (NS) [KP179326]. The total genome size was 13,701 bp. The eight segments of the viral genome encoded 12 proteins (PB2, PB1, PB1-F2, PA, PA-X, HA, NP, NA, M1, M2, NS1, and NS2) with lengths of 759, 757, 90, 716, 252, 566, 498, 469, 252, 97, 230 and 121 amino acids, respectively. After performing the analysis of the HA and NA amino acid sequences from the A/Pekin duck/Mexico/CPA-5009/2007 virus, we identified the virus as the H6N2 subtype.
We found that the nucleotide sequence of each of the eight segments of the A/Pekin duck/Mexico/CPA-5009/2007 virus genome showed 99% similarity with a genetic distance that was closely related to the isolated viruses from wild ducks in North America, corresponding to different subtypes, such as H6N2, H3N8, and H11N2 (Table 1).
Table 1 Increased nucleotide identity in each of the segments of the A/Pekin duck/Mexico/CPA-5009/2007 virus genome.
| Segment | Closer virus | Nucleotide identity (%) | Genetic distance | Access to GenBank |
| (PB2) | A/mallard/Alberta/28/2007(H3N8) | 99 | 0.00313355 | CY103201 |
| (PBI) | A/blue-winged teal/Texas/Sg-00084/2007(H3N8) | 99 | 0.00177556 | CY033729 |
| (PA) | A/mallard/Alberta/27/2007(H3N8) | 99 | 0.00232143 | CY103191 |
| A/mallard/Alberta/28/2007(H3N8) | 0.00232143 | CY103199 | ||
| (HA) | A/American wigeon/California/HKWF371/2007(H6N5) | 99 | 0.0114625 | CY032704 |
| A/northern pintail/California/44241-747/2006(H6N1) | 0.01146808 | FJ520099 | ||
| (NP) | A/mallard/Alberta/76/2006(H1N3) | 99 | 0.00396827 | CY077153 |
| *A/mallard/Alberta/156/2007/(H3,4, N6,8) | 0.00396827 | CY103319 | ||
| (NA) | A/northern shoveler/California/9187/2008(H6N2) | 99 | 0.0057446 | CY094311 |
| (M) | A/blue-winged teal/Louisiana/Sg-00218/2007(H3N8) | 99 | 0.00300677 | CY078234 |
| A/blue-winged teal/Louisiana/Sg-00224/2007(H3N8) | 0.00300677 | CY078354 | ||
| *A/blue-winged teal/Texas/Sg-00082/2007(H3N8) | 0.00300677 | CY077948 | ||
| (NS) | A/gadwall/Mississippi/11OS6084/2011(H11N2) | 99 | 0.00580827 | CY166812 |
| A/northern shoveler/Mississippi/11OS6077/2011(H11N2) | 0.00580827 | CY166804 |
* Genetic distance.
The phylogenetic analysis between the HA of the A/Pekin duck/Mexico/CPA-5009/2007(H6N2) virus, with 100 sequences that shared a similarity higher than 97% with the H6 serotype, showed that the virus was related to the viruses of migratory wild ducks from North America according to the two-parameter Kimura model (Figure 3).
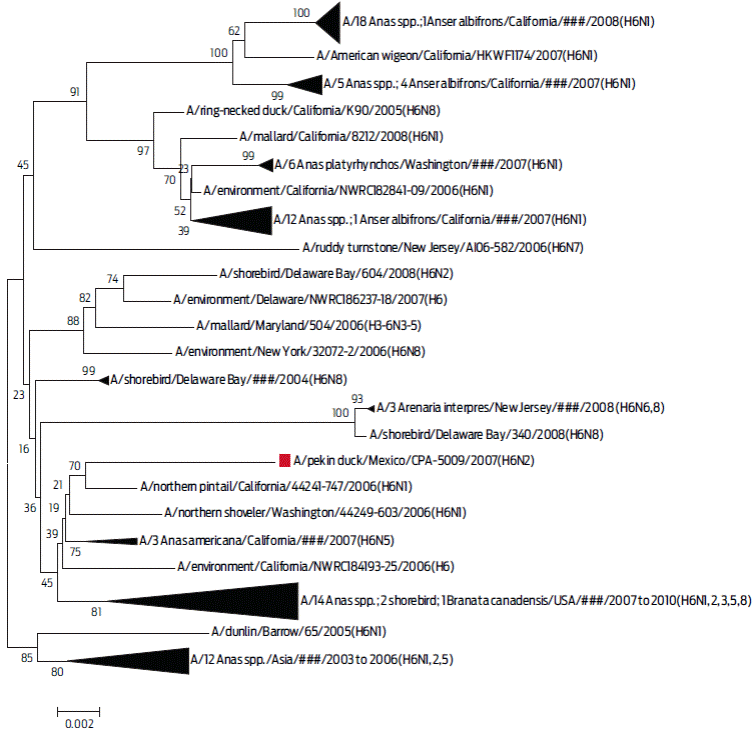
Figure 3 Phylogenetic tree of segment 4 (HA protein). In this dendrogram, the inferred phylogeny is based on the Neighbour-Joining method on 101 selected sequences that had > 97% similarity with the A/Pekin duck/Mexico/CPA-5009/2007 (H6N2) virus. These sequences were included in the phylogenetic analysis of the H6 subtype virus. In each of the nodes, 1000 replica values were observed. The sequence of interest is marked with a red frame. The triangle size reflects the number of nucleotide sequences with a genetic relationship.
The phylogenetic analysis between the NA gene of the A/Pekin duck/Mexico/CPA-5009/2007 virus, with 100 sequences that shared a similarity higher than 98% with the N2 serotype, isolated from the years 2006 to 2013, showed that they are related to viruses from migratory wild ducks in North America according to the two-parameter Kimura model (Figure 4).
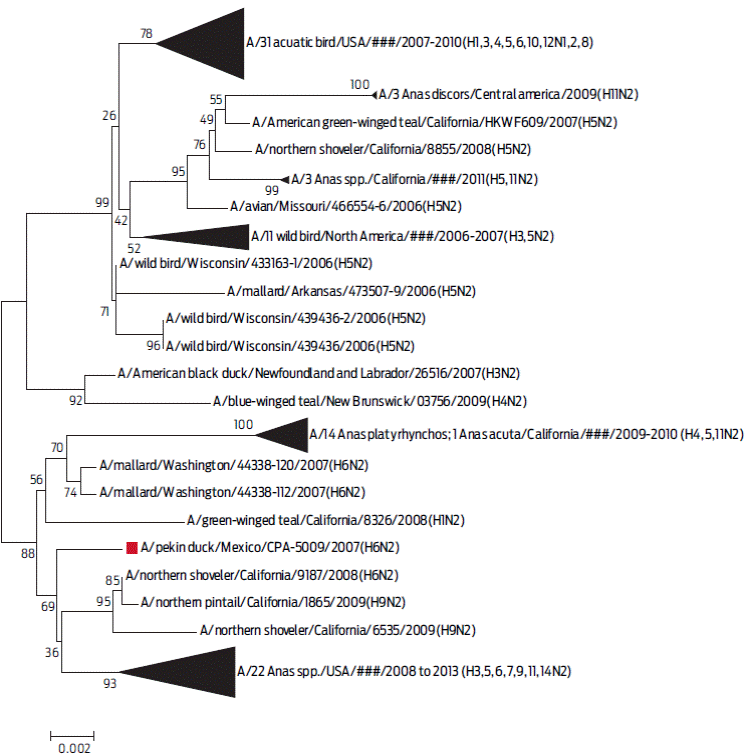
Figure 4 Phylogenetic tree of segment 6 (N protein). In this dendrogram, the inferred phylogeny is based on the Neighbour-Joining method on 101 sequences that had > 97 % similarity with the A/Pekin duck/Mexico/CPA-5009/2007 (H6N2) virus. These sequences were included in the phylogenetic analysis of the N2 subtype virus. In each of the nodes, 1000 replica values were observed. The sequence of interest is marked with a red frame. The triangle size reflects the number of nucleotide sequences with a genetic relationship.
The ≥97% gene similarity of the A/Pekin duck/Mexico/CPA-5009/2007(H6N2) virus genome with the H3N8, H6N1 and H6N2 subtypes suggests previous recombination between different subtypes, as observed in the Alaska Peninsula41 and Southeast Asia.42,43 The high similarity (>97%) among the selected sequences with respect to the gene that codes for the HA and NA genes of the A/Pekin duck/Mexico/CPA-5009/2007(H6N2) virus indicates its relationship to the viruses of migratory ducks from North America that mainly come from the Pacific region. The Pacific region is one of the most important migratory routes that reaches Mexico.
In silico analysis of virus HA, NA, PB1, PB2 and NS1 proteins
The amino acid sequence of the cleavage site between HA1 and HA2 contains PQIETR↓G. Six likely N-glycosylation sites were found in HA1 at 26, 27, 39, 186, 306 and 311 positions; two additional N-glycosylation sites in HA2 at 498 and 557 positions were found. Additionally, there were sialic acid affinity sites in the following HA protein amino acids: F133, T137, and I190 from H6; A225 from H1; and K226 and P228 from H2, H3 and H7. The H6 amino acid sequence showed 99% similarity with the Chinese, Japanese and USA sequences. The NA protein was most similar (99%) to sequences of the North American viruses from the H6N2 and H11N2 subtypes in California and H9N2 in Alaska and Arkansas. The stem region of the protein had a conservative arrangement, maintaining the cysteines of positions 42 and 52, which is characteristic of the N2 protein. The likely N-glycosylation sites in the NA protein were found in the following positions: 61, 69, 86, 146, 200, 234 and 402. NA had E119, H275, R293, and N295 amino acids, which corresponded to the sensitivity determinant sites for the inhibitors of this protein. The NA protein did not have elimination of the stem region and lacked mutations associated with oseltamivir resistance. PB1 maintained an asparagine at position 66, which is associated with low virulence. In PB2, E627K and D701N mutations, which are associated with the interspecies transmission, were not found, while in NS1, the D92 amino acid was associated with non-increased resistance to interferon (IFN).
The glycosylation sites and the PQIETR↓G amino acid sequence at the H6 protein cleavage site of the A/Pekin duck/Mexico/CPA-5009/2007 (H6N2) virus correspond to the previously reported characteristics of these subtypes.42,44 The HA protein of the A/Pekin duck/Mexico/CPA-5009/2007 (H6N2) virus can bind to α2-3 sialic acid present in birds without demonstrating mutations that adapt to human receptors by binding to α2-6.45 In addition, an absence of aspartic acid insertions between the 144 and 145 positions of the HA protein was observed, indicating that the virus originates from aquatic birds.44
The presence of the complete stem region of the NA protein from the A/Pekin duck/Mexico/CPA-5009/2007 (H6N2) virus demonstrates that the virus has not adapted to poultry.46 No mutations were found in association with resistance to antivirals, such as oseltamivir, indicating that the virus is sensitive to NA inhibitors.47 PB1 did not show any mutations associated with virulence, which is the case in mice and ferrets.48 In PB2, we did not find any associated mutations with interspecies transmission,49 whereas NS1 did not have changes in the amino acids that increase IFN resistance, which is in contrast with humans.50
The H6 subtype of viruses, more than any other subtype, infects a wide variety of hosts, suggesting that they can rearrange with other viruses, cross the species barrier and infect mammals, including humans, without adaptation.1 The H6N2 subtype of influenza viruses can infect poultry,51 but little is known about its frequency in these birds.52 In this study, a link between H6N2 in duck and poultry was not found.
Ethical considerations
The study was approved by the Committee for Animal Experiments at the Faculty of Veterinary Medicine and Animal Husbandry (SICUAE), number MC-2016/2-1.
Conclusion
This study found that a temporary, small artificial pond used for human activities (agriculture irrigation, fish and backyard livestock as cattle, swine and poultry) with up to 9000 wild migratory ducks from nine different species during the wintering season may provide a suitable biological setting to support the presence of IAV, but the minimum biological environment to isolate IAV is still unknown. The fact that a single IAV was isolated and identified in this study represents a lower rate of detection than that found in other studies with sentinel ducks. Future surveillance programs need to include waterfowl populations and richness measurements, as well as sampling of the sediment and water to better understand IAV transmission in specific wetlands over time and space. As far as we know, this is the first H6N2 subtype and the third IAV isolated from wildlife in Mexico.











 nueva página del texto (beta)
nueva página del texto (beta)