Introduction
Infectious diseases of poultry, especially bacterial diseases of the respiratory sys tem, cause significant losses in poultry production due to the increased mortality of birds and high treatment costs.1 Over the last 10 years, the poultry industry in Sonora noted a considerable decrease in egg production. It was established that this decrease was associated with Pasteurella haemolytica-like bacteria, but since these bacteria were considered normal gut flora, this line of investigation was not pursued further.2 Some years later, genetic changes in these bacteria were associ ated with an increase in virulence. This discovery led to the establishment of a new genus within the family Pasteurellaceae, and the classification of these bacteria as Gallibacterium anatis biovar haemolytica.3
Gallibacterium anatis is common in microbiological analyses and affects both the respiratory and reproductive systems. Infection may cause a decrease in egg production and produce a variety of lesions, including follicular atrophy, oophoritis, septicaemia, misshapen follicles, peritonitis, enlarged kidneys, enteritis, congestion, and oedema of the respiratory tract.2,4,5 Mirle et al.6 established that Gallibacteri um anatis is also associated with variation in egg size, eggshell formation, and re ductions of 4 % to 20 % in egg production. Gallibacterium anatis represents both a significant health risk and an economic challenge for the Mexican poultry industry.
In the present study, Gallibacterium anatis isolates were isolated from com mercial layer hens in the southern part of the state of Sonora to determine the biovars in the region. These results will be used to implement improved control programmes based on the use of the correct immunogens. Classification of Galli bacterium anatis can be based on biochemical reactions.7 Thirteen biovars of G. anatis, 7 of which have been reported in Mexico (1, 2, 4, 5, 8, 16 and U), can be identified using the approach developed by Jaworski et al.7 Bisggard developed a biochemical classification system with 24 biovars, thereby providing a more specific classification system.8,9 Based on the latter classification system, biovars 1, 4, 11, 12, 13, and 22 are present in Mexico, with occasional reports of biovars 3, 7, 15, 16, 17, 18, 19, 20, and 24. Biovars 1 and 4 have been reported in the southern areas of the state of Sonora.2,3
Material and methods
Sampling
A total of 600 hens, twenty birds each from 30 farms, with respiratory and repro ductive signs and medical histories were collected. Sampling and necropsies were performed following the procedures described elsewhere.10,11 This study was car ried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and all efforts were made to minimize suffering.
Isolation and identification of Gallibacterium anatis
Swab samples collected from the turbinates, trachea, lungs, cleft palate, liver, spleen, kidneys, follicles, and peritoneum were cultivated on blood agar (BD Bi oxon) containing 5 % sterile defibrinated bovine blood and on MacConkey agar (BD Bioxon). The samples were incubated at 37 °C for 18 to 24 hours in a microbiological incubator (Shel Lab 1535).12,13 For morphological identification of bacteria, colonies with a diameter of 1 to 2 mm that were circular, convex, and translucent grey, were purified and selected based on Christensen’s criteria.3 Bacteria were stained with Gram stain, visualized using an Olympus microscope CX41, and identified as gram-negative coccobacilli.14-16 Biochemical procedures for the identification of bacteria were then performed using cultures that were < 24 hours old, and carbohydrate fermentation, enzyme production, and the production of certain metabolites were analysed using MicroScan Renok™ for gram-negative bacteria. Following the manufacturer’s recommended procedure, inoculated plates were incubated at 37 °C for 24 hours (Shel Lab 1535). Because of the absence of Gallibacterium anatis in the MicroScan database, manual interpretation was used, i.e. in each well, a specific colour change was evaluated as a surrogate for a meta bolic reaction. In addition, biochemical analyses for other carbohydrates (glucose, maltose, trehalose and D-xylose) were performed (BD Bioxon) to complement the biochemical tests and identify the bacteria. All biochemical tests were performed twice. In addition, the oxidase test was performed for the presence of the oxidase enzyme using a dipstick (Bactident® Oxidase). Isolate typing was based on pre vious reports of the carbohydrate fermentation of (+) L-arabinose, (+) D-xylose, m-inositol, (-) D-sorbitol, maltose, trehalose, and dextrin, that differentiated the 24 registered biovars (Table 1).3,8
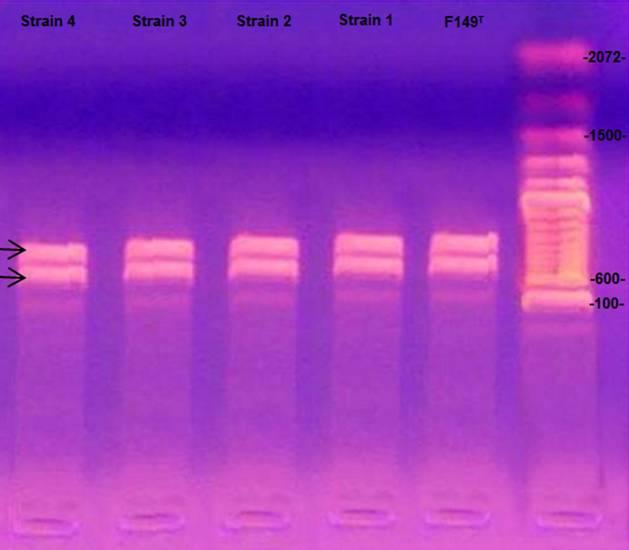
Thereafter, to confirm bacterial identity, polymerase chain reactions PCR tar geted to amplify specific gene segments of Gallibacterium anatis were performed. Bacterial DNA was extracted using a QIAamp DNA Mini Kit (51306, QIAGEN) follow ing the manufacturer’s instructions. The DNA used in the (PCR) was adjusted to a concentration of 50 μg/μl. The primers and oligonucleotides were designed based on the 99 sequences of 16S rRNA from Gen Bank.17 The primer 1133fgal (5’-TAT TCTTTGTTACCARCGG) is specific to Gallibacterium and does not amplify DNA from other Pasteurellaceae within the chosen PCR conditions. For inverse amplification, the primer of the 23S rRNA gene with the sequence 114r (5’-GGTTTCCCCATTCGG) was selected.18,19 PCR was performed with the default settings of the thermocycler (ThermoFisher Gene 9700) and with 2.5 µl of Mg 10X buffer, 0.75 µl of MgCl2 (50 mM), 0.5 µl of dNTPs mix (10 mM), 0.25 µl each of F primer and R primer (25 µM), 0.25 µl of Taq (5 U/µl), 18.5 µl of RNase-free H2O, and 2 µl of DNA from each of the samples. Denaturation was performed for 4 minutes at 95 °C, followed by 35 cycles of 95 °C for 30 seconds each, and then the alignment was performed for 1 min at 54 °C and 2 min at 72 °C, followed by a final extension of 72 °C for 10 min. The amplification products (790 bp and 1080 bp for the genus Gallibacterium) were examined by the separation of PCR products during electro phoresis on 1.6 % agarose gel stained with ethidium bromide.18,20 The amplicons were visualized on a UV transilluminator (UV-25 Macro Vue, Hoefer).
Results and discussion
In this study, a known isolate from previous studies was included as a control. Iso lates of Gallibacterium spp. were identified by their morphological and haemolytic characteristics as described by Christensen et al.3 Although 600 samples were analysed, the recovery of Gallibacterium spp. was not possible in all cases, per haps because hens were previously immunized against this organism. These results support the idea that this organism causes respiratory and reproductive disease because 60 % of the isolates of Gallibacterium anatis were obtained from lungs: 30 % in tracheae and 30 % in follicles.
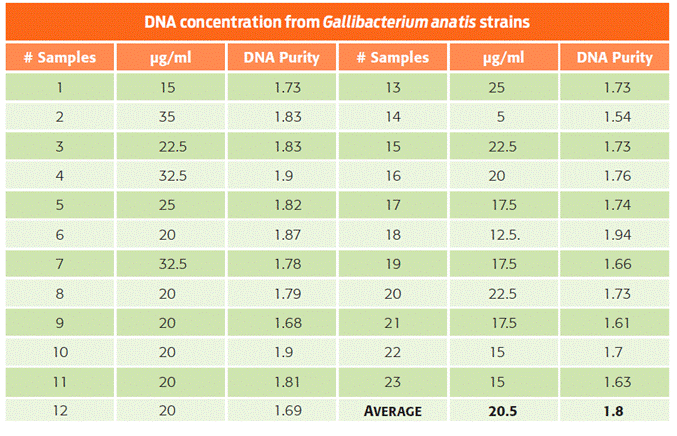
The classification of biovars via biochemical tests3,8,9 showed that 41 % of the bacteria were biovar 1, and 59 % were biovar 6 (Table 1). As opposed to previous reports, biovar 4 was not recovered in this study.2 In the extraction process, an average of 20.5 μg/μl of DNA and a DNA purity ratio of 1.8 were achieved (Table 2).
Table 2 Concentration and purity of the extracted DNA from each bacterial isolate of Gallibacterium anatis.
Likewise, the presence of Gallibacterium anatis was confirmed by G. anatis-specific PCR for 16S rRNA and 23S rRNA.19 As reported elsewhere,3,19,21 these procedures rule out other members of the family Pasteurellaceae. The amplification products of the isolated bacteria were 790 bp and 1080 bp long, thereby confirming the identity of the 23 isolates as Gallibacterium anatis (Figure 1).
Diagnostics performed in the state of Sonora between 2009 and 2011 identi fied biovar 6 and biovar 1 in birds immunized with commercial products containing biovar 1 and biovar 4. These findings suggested that the outbreak might have occurred because biovar 6 was absent in the commercial immunogen.2 Based on these results, it is recommended that immunogens include biovar 6 to prevent future outbreaks.
The identification of strains of G. anatis is often burdensome because of the difficulty in interpreting phenotypic data.3,8 For this reason, PCR is highly useful in the diagnosis of Gallibacterium anatis. Although the use of molecular methods is constrained by the high cost of PCR, biotyping is an important tool in outbreak investigations. Tegtmeier et al.21 demonstrated that bacterial identification through conventional methods is less sensitive than either immunohistochemistry or PCR. This highlights the importance of having alternative diagnostic tools available for G. anatis in order to rule out other gram-negative pathogens associated with similar clinical signs, e.g., E. coli, and start specific treatment.8,19
Conclusions
Overall, the results of this study showed the presence of Gallibacterium anatis in association with respiratory and reproductive problems in populations of com mercial laying hens in southern Sonora. Additionally, for the first time in Mexico, G. anatis biovar 6 was detected. The presence of this new biovar will provide a basis for understanding pathogenesis and immunity, and it may explain the clinical presentation of Gallibacterium anatis in immunized birds.











 nueva página del texto (beta)
nueva página del texto (beta)