Introduction
Paratuberculosis (PTB) is an infectious disease characterized by diarrhea and granulomatous enterocolitis in domestic and wild ruminants. Mycobacterium avium subsp. paratuberculosis (MAP) is the etiological agent of PTB. This pathogen is capable of infecting enteric macrophages and lives intracellularly. In order for bacteria to invade and multiply within host cells, the cells must meet specific metabolic requirements. A key element among them is iron, a cofactor of many cellular proteins that participates in electron transport, ROS detoxification and DNA synthesis (Eckelt et al., 2014, 2015). Iron is not freely available; therefore, bacteria utilize various iron uptake mechanisms to acquire it. Production of siderophores, hemophores and cell surface receptors that catch hemoproteins helps bacteria to circumvent this problem (Krewulak and Vogel, 2008). In mycobacteria, mycobactin is a siderophore responsible for the binding of iron and its transport into cells. Mycobactin is encoded by a gene cluster designated mbtA-J; in MAP, mbtA is shorter and encodes a truncated version of the protein that initiates mycobactin synthesis. Truncation of mbtA may explain why MAP is not able to produce mycobactin in a laboratory culture (Li et al., 2005). Despite the in vitro requirement for mycobactin, MAP overexpresses the genes involved in mycobactin synthesis in bovine macrophages (Zhu et al., 2008). In the presence of iron, the MAP transcriptional regulator ldeR recognizes consensus sequences in “iron boxes” of promoter sequences and regulates the expression of genes involved in iron acquisition (mbt) and storage (bfrA) (Janagama et al., 2009, 2010). The interactome of another iron acquisition mechanism dependent on the presence of nitric oxide has been recently characterized in MAP (Lamont et al., 2013).
The regulation of iron homeostasis in macrophages involves numerous proteins (Weiss, 2009). Macrophages can acquire iron through transferrin receptor 1 (Tfr1) and the transferrin-mediated capture of iron. Molecular iron can be obtained through transporter 1 (Dmt1) or by phagocytosis of old erythrocytes. Iron is then transported, reused and stored in macrophages through the interaction of iron regulatory elements (IREs) with iron regulatory proteins (IRPs). Ferroportin 1 (FPN1) is an iron export protein that is present on the surface of macrophages, hepatocytes, enterocytes, and erythrocytes. Ferroporitn 1 is specific for the ferrous form of iron (Fe2+) (Abboud and Haile, 2000; Donovan et al., 2000; McKie et al., 2000). Iron efflux is controlled in macrophages by hepcidin, which interacts with FPN1. This interaction results in FPN1 internalization and degradation by proteasomes, which blocks iron efflux in macrophages (Nemeth et al., 2004). Accumulation of iron inside the cell promotes the development of intracellular pathogens and decreases the secretion of IL-6 (Ganz, 2009). Macrophage FPN1 mRNA expression is increased by Mycobacterium tuberculosis (MTB) and Mycobacterium avium (MA) infection. However, an even higher level of increase of fpn1 gene expression in MTB- and MA-infected macrophages was achieved when macrophages were classically activated by interferon gamma (IFNγ) (Van Zandt et al., 2008). The effect of MAP on FPN1 mRNA expression is not well understood.
Considering the roles and relevance of FPN1 and iron in macrophages during MAP infection, we hypothesized that a high concentration of iron is necessary for the survival of mycobacteria in macrophages during the process of infection. Consequently, the FPN1 mRNA concentration should be decreased to facilitate the establishment of infection. In this study, we aimed to examine FPN1 mRNA expression levels via real-time polymerase chain reaction (PCR) in the mouse macrophage cell line J774 that was incubated in the presence of live MAP, with or without an iron overload treatment, and MAP crude protein extract. Our results identified the down-regulation of macrophage FPN1 mRNA expression as a direct effect of MAP infection.
Materials and methods
Bacterial cultures and crude protein extract
MAP (ATCC 19698) was cultivated in Middlebrook 7H9 medium supplemented with oleic acid, albumin, dextrose and catalase (OADC) (Difco, Detroit, MI, USA) and mycobactin (Allied Monitor Inc., USA)(4 mg/L) for 4 weeks. Bacteria were stored at -70°C in 1-mL aliquots with 4 x 108 colony forming units (CFU)/mL in RPMI 1640 medium (Difco, Detroit, MI, USA) containing L-glutamine, amino acids, sodium pyruvate and sodium bicarbonate (complete RPMI [CRPMI]) with 10% glycerol. The number of bacteria was confirmed by diluting the samples and plating them on Middlebrook 7H11 agar supplemented with OADC and mycobactin. Mycobacterial crude protein extracts were prepared from 50 mL of culture after 1 month of growth. The bacterial suspension was centrifuged for 20 min at 3,000 X g; the pellet was washed with PBS and resuspended in CRPMI. The suspension was disrupted in a bread beater in a proportion of 1 g/mL for 5 min and centrifuged for 20 min at 3,000 X g to eliminate cellular debris. The supernatant was filtered through a 0.22 µm membrane. The protein concentration of the filtered extract was determined by the Bradford method. Aliquots of crude protein extract were kept at -70°C until further use.
Cell culture of J774 murine macrophages
The murine macrophage cell line J774 was propagated in CRPMI enriched with 5% heat-inactivated fetal bovine serum without antibiotics. Cultures were maintained at 37°C with 5% CO2. Macrophages were quantified in a Neubauer chamber after dilution with trypan blue. Six-well culture plates were then seeded with 1 x 106 cells per well, and incubated at 37°C with 5% CO2 for 4 hours to allow adhesion to the surface. Non-adherent cells were removed by washing with CRPMI supplemented with 5% fetal bovine serum. Adherent macrophages were then used for subsequent experiments.
Macrophage treatment with live MAP and crude protein extract
In an attempt to identify the effect of live MAP and crude protein extract on macrophage fpn1 gene expression, macrophage monolayers containing 1 x 106 cells in tissue culture plates were infected with either live MAP cells or treated with MAP crude protein extract. For the infection, 4 different multiplicities of infection (MOIs 5:1, 10:1, 15:1 and 20:1) were added to the wells, and then the plates were centrifuged at 200 X g for 10 min and incubated at 37°C for 6 hours. For the protein treatment, wells were incubated with 50, 100, 150 or 200 µg/mL of MAP crude protein extract for 6 hours. At the end of the incubation, culture medium was removed from all treatments, cells were washed once with PBS, and total RNA was extracted. Macrophage fpn1 gene expression results are the average of 3 independent experiments each one with 3 replicates.
Macrophage treatment with ferric nitrilotriacetate (FeNTA)
To identify the effect of an iron overload treatment on macrophage fpn1 gene expression, macrophages were incubated with different concentrations of ferric nitrilotriacetate (FeNTA), a transferrin iron donor. J774 murine macrophages (1 x 106 cells) were treated with 100, 200 and 400 µM FeNTA for 6, 9 or 12 hours in CRPMI medium supplemented with 5% fetal bovine serum. In an attempt to determine the effect of MAP infection in macrophages with an iron overload, macrophages were either MAP-infected (MOI 20:1), treated with 200 μg/mL of MAP crude protein extract, treated with 400 μM FeNTA, or MAP-infected (MOI 20:1) in the presence of 400 μM FeNTA (FeNTA-MAP). For all treatments, the medium was removed at the end of the incubation time, cells were washed once with PBS, and total RNA was extracted and stored at -70°C. Macrophage fpn1 gene expression results were obtained from 3 independent experiments with 3 replicates each.
RNA extraction
Macrophage total RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations and instructions. All samples were treated with DNase (RNase-Free DNase Set) and purified using columns from the RNeasy MinEluteTM Clean kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. RNA integrity was assessed by 1% agarose gel electrophoresis, while quantification was determined by measuring the 260/280 nm optical density ratio using a NanoDrop (Thermo Fisher Scientific Inc., Waltham, MA, USA).
cDNA synthesis
Following the manufacturer’s instructions, cDNA was synthesized using 4 U of the reverse transcriptase enzyme Omniscript1 (Qiagen, Hilden, Germany) in a final volume of 20 µL containing an RT buffer (1x), dNTPs (0.5 mM each one), oligo-dT primers (1 µM), total RNA template (1 µg) and nuclease-free water. The mixture was incubated for 60 min at 37°C, and the enzyme was inactivated at 95°C for 5 min. The final volume was adjusted to 400 µL with nuclease-free water and stored at -80°C until use.
Real-time PCR
Primers specific for fpn1 gene (Forward 5´ GCGTCATTGCTGCTAGAATCG 3´, Reverse 5´ CATGGAGTTCTGCACACCGTT 3´) and for the housekeeping gapdh gene (Forward 5´ ACGGCACAGTCAAGGCAGAGAA 3´, Reverse 5´ TCTCGCTCCTGGAAGATGGTGA 3´), which was used as an internal control, were designed using the Primer express 2.0 software. Real-time PCR was performed using a Roche LightCycler® 480 II (Roche, Indianapolis, IN, USA) and a Roche LightCycler® 480 DNA SYBR Green I Master reagent (Roche, Indianapolis, IN, USA). Amplification reactions were performed in a final volume of 20 µL containing 10 µL of 2X SYBR Green I master reagent buffer, primers (F and R, 0.1 μL each), 5 µL of diluted cDNA and 4.8 µL of nuclease free water. All reactions were run in 96-well optical plates with an initial incubation of 95˚C for 10 min, and then 45 cycles of 95˚C for 10 s, 60˚C for 15 s and 72˚C for 15 s during which the fluorescence data were collected. At the end of the 45 cycles, a final extension was run for 5 min at 72˚C. For each target, a melting curve analysis was performed to demonstrate specificity, and the efficiency of the PCR amplification was determined by assaying the amplification of serially diluted cDNA samples. A negative control without cDNA (NTC, No Template Control) was run in each plate to assess the overall specificity; each sample was set up in triplicate. A threshold cycle (Ct) was determined for each sample by examining exponential growth phase and the baseline signal from fluorescence versus cycle number plots. A sample was deemed positive if fluorescence exceeded the threshold. Melting curves were transformed to negative first derivative melting curves. In the melting curve analysis, the negative first derivative peaks, which are characteristic of the PCR product melting temperature, were used to identify specific PCR products. Amplification reactions were also routinely checked for the presence of nonspecific products by agarose gel electrophoresis. Threshold cycle values of the sample were used to calculate the units of relative expression (URE) by the comparative 2 -ΔΔCt method (Qiagen integrative solutions - real-time PCR applications, 2006). The results were obtained from 3 independent experiments with 3 replicates each.
Results
MAP infection of murine macrophages induced differential FPN1 mRNA expression dependent on the multiplicity of infection
Different MOIs of MAP were used in this experiment to determine a MAP dose-response in J774 murine macrophages. Infection with live MAP decreased FPN1 mRNA levels in an MOI dependent fashion. In comparison with the control group, macrophages infected with MOIs of 20:1 and 15:1 did not show a significant change in fpn1 gene expression, whereas MOIs of 10:1 and 5:1 induced a decrease of 50 and 80%, respectively (P ≤ 0.05, Figure 1).
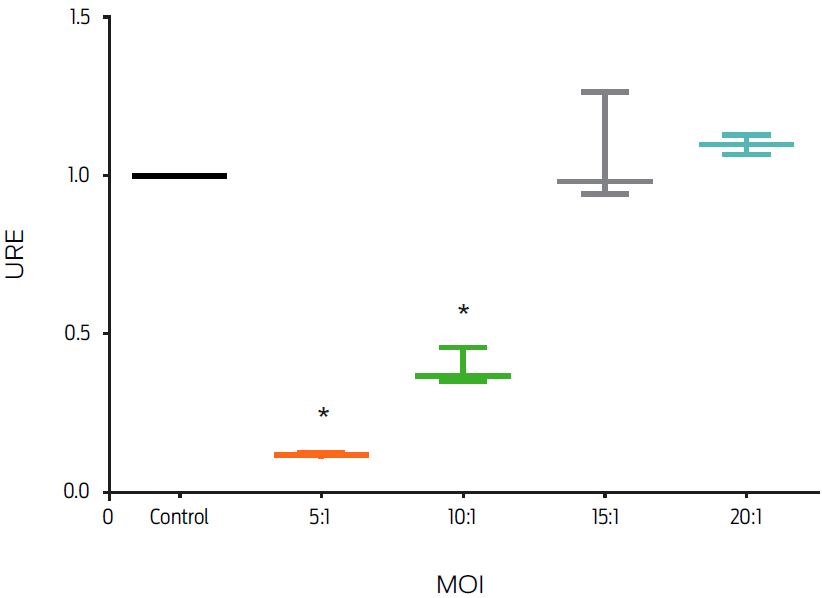
Figure 1: Infection by Mycobacterium avium subsp. paratuberculosis (MAP) down-regulates macrophage fpn1 gene expression. Murine macrophages, J774, were infected with MAP at different multiplicities of infection (MOIs) for 6 hours. Total RNA was extracted and gene expression was measured by real-time PCR. Results were normalized to GAPDH and expressed as units of relative expression (URE). Results are the mean ± standard deviation of 3 independent experiments with 3 replicates each. Statistical significance, P ≤ 0.05.
Stimulation with different concentrations of MAP crude protein extract decreased the FPN1 mRNA expression in murine macrophages
The effect of MAP crude protein extract on FPN1 mRNA expression was analyzed. In this study, we decided to use a protein concentration range that covered the limits of the experimental variation previously reported (Ciaramella et al., 2004; López et al., 2003, 2015; Sanchez et al., 2008). Macrophages treated with 50, 100, 150 and 200 µg/mL of MAP crude extract decreased FPN1 mRNA expression by 25% (P ≤ 0.05, Figure 2).
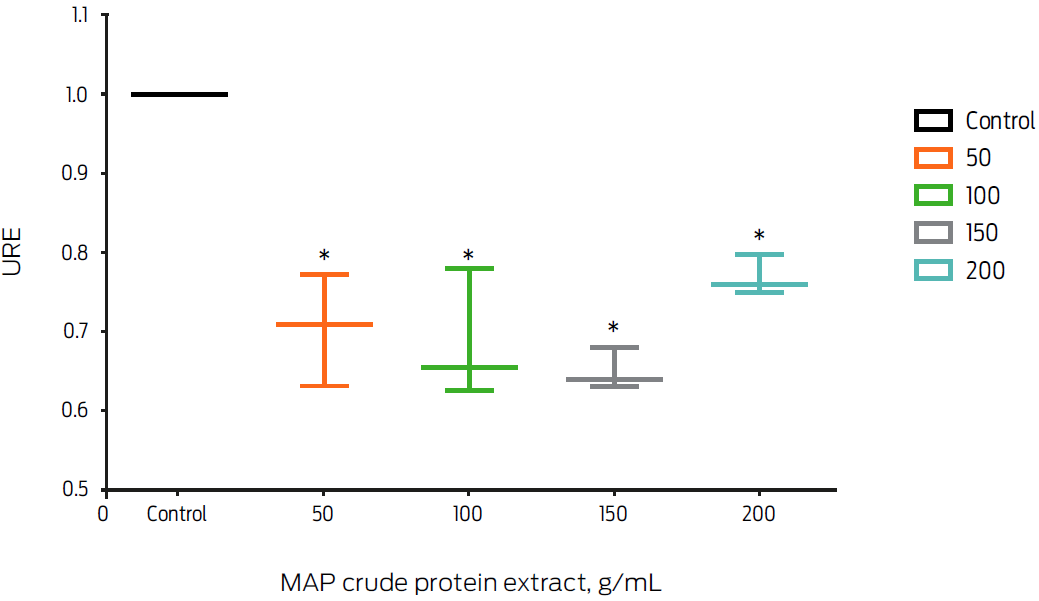
Figure 2: Macrophage treatment with Mycobacterium avium subsp. paratuberculosis (MAP) crude protein extract slightly down-regulates fpn1 gene expression. Murine macrophages, J774, were incubated with different concentrations of MAP crude protein extract for 6 hours. Total RNA was extracted and gene expression was measured by real-time PCR. Results were normalized to GAPDH and expressed as units of relative expression (URE). Results are the mean ± standard deviation of 3 independent experiments with 3 replicates each. Statistical significance, P ≤ 0.05.
Treatment with different concentrations of FeNTA induced FPN1 mRNA expression in murine macrophages
Induction of FPN1 mRNA expression by iron overload was measured in the presence of 100, 200 and 400 μM of the transferrin iron donor FeNTA for 6, 9 and 12 hours. Macrophages treated with FeNTA showed an increase in fpn1 gene expression at all of the analyzed time points (P ≤ 0.05, Figure 3). The increase in gene expression was FeNTA concentration-dependent. However, the treatment using 400 μM at 6 hours induced the highest increase (10-fold). Therefore, these conditions were used in subsequent experiments.
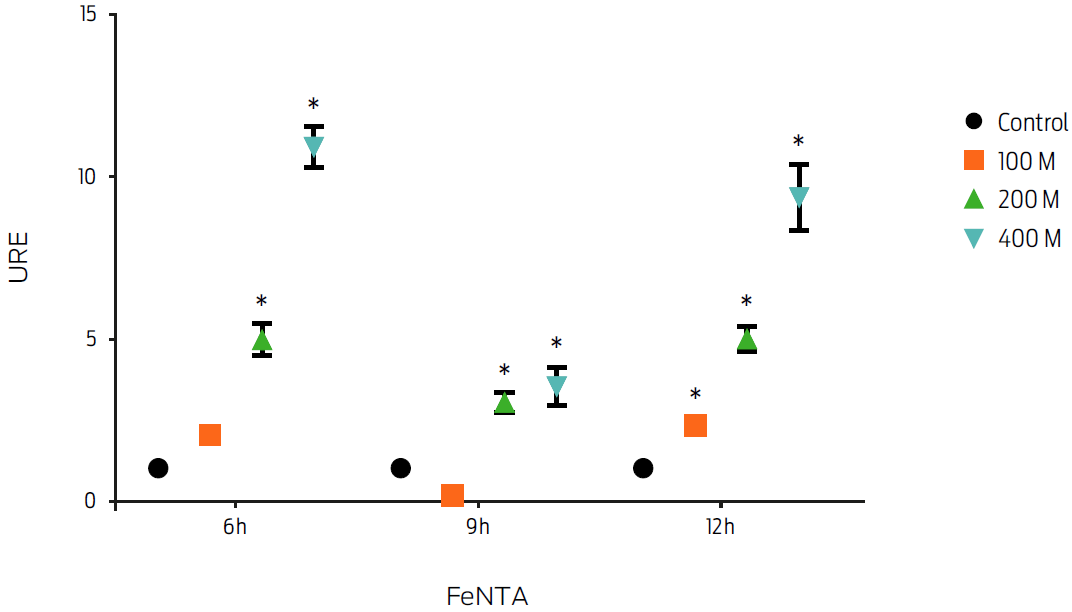
Figure 3: Macrophage treatment with ferric nitrilotriacetate (FeNTA) increases fpn1 gene expression in a dose dependent manner. Murine macrophages, J774, were incubated with different concentrations of FeNTA for 6, 9 and 12 hours. Total RNA was extracted and gene expression was measured by real-time PCR. Results were normalized to GAPDH and expressed as units of relative expression (URE). Results are the mean ± standard deviation of 3 independent experiments with 3 replicates each. Statistical significance, P ≤ 0.05.
FPN1 mRNA expression induced by iron overload (FeNTA) is down-regulated by MAP infection in J774 murine macrophages
The effect of MAP infection on FPN1 mRNA expression in macrophages with an iron overload was measured. MAP-infected macrophages incubated with 400 μM FeNTA (FeNTA-MAP) exhibited a 70% down-regulation of fpn1 gene expression compared to the gene expression in uninfected, FeNTA-treated macrophages (P < 0.05, Figure 4).
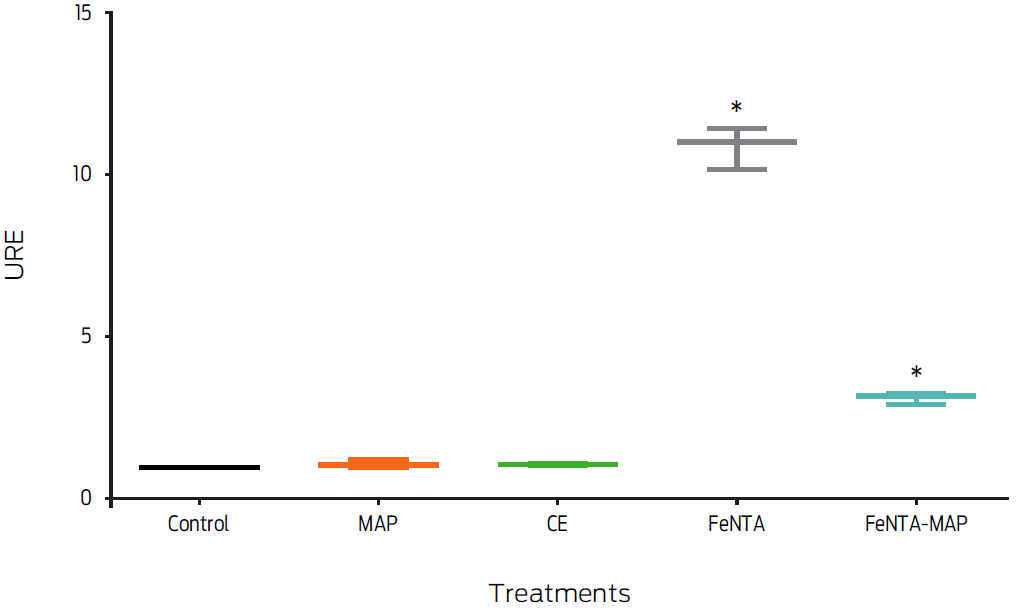
Figure 4: Macrophage treatment with ferric nitrilotriacetate and Mycobacterium avium subsp. paratuberculosis (FeNTA-MAP) decreases fpn1 gene expression. Murine macrophages, J774, were treated with 400 µM of FeNTA, 200 mg/mL of crude extract (CE), a MOI of 20:1 MAP or with 400 µM of FeNTA and a MOI of 20:1 MAP (FeNTA-MAP) for 6 hours. Total RNA was extracted and gene expression was measured by real-time PCR. Results were normalized to GAPDH and expressed as units of relative expression (URE). Results are the mean ± standard deviation of three independent experiments with three replicates each. Statistical significance, P ≤ 0.05.
Discussion
Ferroportin 1 mRNA expression in J774 mouse macrophages was down-regulated following treatment with live MAP or its crude protein extract. Down-regulation induced by viable bacteria was greater, suggesting that the metabolic activity of the bacteria played a role in the process. Differential fpn1 gene expression was MOI-dependent. MOIs of 10:1 and 5:1 induced a decrease of 50 and 80%, respectively, whereas 20:1 and 15:1 did not modify the mRNA levels. Our results are controversial; however, we may argue that the amount of bacteria infecting a macrophage monolayer may generate different results depending on the multiplicity of infection. The interaction of bacterial PAMPs with macrophage receptors can stimulate a variety of responses; however, under a high concentration of bacteria, saturation of the receptor may stop or change the direction of these responses. Furthermore, an increase in stimulation may activate other signaling pathways that interfere with responses elicited under a low bacterial concentration. For instance, Mycobacterium tuberculosis induces macrophage apoptosis with the participation of caspases when MOIs ≤ 10:1 are applied, but caspase activation is turned off at an MOI of 25:1 (Lee et al., 2006). On the other hand, macrophages infected with a high bacterial burden may try to redirect gene expression in an attempt to survive. It is reasonable then to propose that a shift in cellular conditions may modify the cell response scenario. Our results note that macrophages infected with an MOI of 20:1 were able to decrease fpn1 gene expression when the cell suffered an iron overload. This result further supports our experimental evidence, which shows a decrease in FPN1 mRNA levels as a direct effect of MAP.
There is evidence that fpn1 gene expression is modified by bacterial infection. For instance, down-regulation of FPN1 mRNA expression in monocytes and macrophages has been linked to exposure to Gram-negative pathogens and LPS, as part of the acute inflammatory response where IFNγ and Toll-like receptors are also key players (Yang et al., 2002; Ludwiczek et al., 2003; Peyssonnaux et al., 2006). In contrast, MTB and MA up-regulated FPN1 mRNA levels in the mouse macrophage cell lines RAW264.7 and AMJ2-C8 under IFNγ activation (Van Zandt et al., 2008). However, in the absence of IFNγ, MTB and Mycobacterium bovis BCG down-regulated fpn1 gene expression in a time dependent manner (Li et al., 2013). All together, these observations suggest a role for bacteria in the transcriptional regulation of FPN1, which is also being regulated by the immune response in an attempt to push the balance in favor of the host. It is important to note that our experiments did not address the effect of MAP in the presence of immune mediators such as LPS or IFNγ. The influence of these molecules on FPN1 mRNA expression in MAP-infected macrophages may modify our findings.
Host cells and pathogens compete for intracellular iron (Weiss et al., 1994). From the pathogen’s point of view, a decrease in FPN1 is beneficial, maintaining the intracellular iron levels necessary for bacterial survival; however, from the host’s point of view, an increase of FPN1 allows the iron to be transported away from intracellular bacteria (Janagama et al., 2009). Our results demonstrated that MAP infection decreases fpn1 gene expression under several different conditions. This observation indicates that MAP induces an effect that has been previously documented with other species of mycobacteria. In terms of virulence, this could be a strategy to support bacterial survival.
Conclusions
We demonstrated that live MAP or its crude protein extract decreased FPN1 mRNA levels. Our data revealed an inhibitory effect of live MAP on FPN1 mRNA and suggested a bacterial mechanism that may play a role in host iron regulation, thus helping bacteria to endure the hostile macrophage microenvironment.











 texto en
texto en 



