Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Abanico veterinario
versión On-line ISSN 2448-6132versión impresa ISSN 2007-428X
Abanico vet vol.10 Tepic ene./dic. 2020 Epub 02-Mar-2021
https://doi.org/10.21929/abavet2020.15
Literature Review
Metabolism in ruminants and its association with blood biochemical analytes
1Estudiante de Maestría en Ciencias Agropecuarias, Universidad Autónoma Metropolitana. México.
2Departamento de Producción Agrícola y Animal, Universidad Autónoma Metropolitana. México.
3Facultad de Medicina Veterinaria y Zootecnia, Universidad de Colima. México.
The present study is an analysis of scientific elements on the metabolism of ruminants: polysaccharides, proteins and lipids. Where i) the fermentative digestion carried out by microorganisms, ii) the posruminal digestion and absorption and iii) the metabolism of each monomer is associated with the blood analytes that give us an approximation to the nutritional metabolism of the animal, also confer information on alterations and adjustments homeostatic. This review emphasizes the metabolism of monosaccharides, amino acids, and fatty acids. Therefore, the revised information aims to make the understanding of catabolic and anabolic processes in ruminant nutrition.
Keywords: glucose; lipids; polysaccharides; proteins and urea
El presente estudio es un análisis de elementos científicos sobre el metabolismo de los rumiantes: polisacáridos, proteínas y lípidos. Donde i) la digestión fermentativa realizada por microorganismos, ii) la digestión y absorción posruminal y iii) el metabolismo de cada monómero, se asocian con analitos sanguíneos que otorgan una aproximación al metabolismo nutricional del animal, además confieren información sobre alteraciones y ajustes homeostáticos. Esta revisión hace énfasis en el metabolismo de monosacáridos, aminoácidos y ácidos grasos. Por lo tanto, la información revisada pretende hacer más accesibles los procesos catabólicos y anabólicos en la nutrición de los rumiantes.
Palabras claves: glucosa; lípidos; polisacáridos; proteínas y urea
INTRODUCTION
Mammals classified as ruminants are characterized by the morphophysiological adaptation of their digestive system (Resende Jr et al., 2019; Rotta et al., 2014), divided into four chambers: I) reticulum, II) rumen, III) omasum and IV) abomasum (Qiyu et al., 2019). Abomasum secretes digestive hydrolases and its function is similar to that of monogastric stomachs (Agarwal et al., 2015). Ruminants specialize in their ability to feed on pasture and forage (Puppel y Kuczyńska, 2016), as they can degrade structural polysaccharides for example: cellulose, hemicellulose and pectin (DePeters y George, 2014), very poorly digestible for non-ruminant species (Kittelmann et al., 2013; Zeng et al., 2017). Food degradation is mainly carried out by fermentative digestion, carried out by microorganisms present in the rumen (Ginane et al., 2015; Wallace et al., 2017). The molecules resulting from ruminal fermentation are used to satisfy the animal's physiological processes (Kittelmann et al., 2013; Li et al., 2019a). The quantification of biochemical analytes in plasma and/or serum, provide an approximation to nutritional metabolism (García et al., 2015). They also confer information on homeostatic alterations and adjustments (Moyano et al., 2018). For this reason, it is important to understand the catabolism and anabolism processes that are carried out in the ruminant to understand the levels of analytes present (Puppel y Kuczyńska, 2016). Because of this, it is necessary to increase our understanding of the metabolism of monosaccharides, amino acids (aa) and fatty acids. Therefore, a bibliographic review was carried out on its metabolism in ruminants and its association with different biochemical analytes.
Abbreviations
aa |
amino acids |
AcAc |
acetoacetate |
AGNE |
unesterified fatty acids |
AGV |
volatile fatty acids |
ALB |
albumin |
Arg |
arginine |
C=O |
carbonyl group |
C16:0 |
palmitic |
C3H3O |
pyruvate |
C6H12O6 |
glucose |
CO2 |
carbon dioxide |
COL |
cholesterol |
COOH |
carboxyl group |
CH4 |
methane |
FAD |
flavin-adenine dinucleotide |
Glu |
glutamic |
H2CO3 |
carbonic |
HCl |
Hydrochloric |
HCO3 |
hydrogencarbonate anion |
His |
histidine |
Ile |
isoleucine |
K+ |
potassium ion |
Leu |
leucine |
Lys |
lysine |
Met |
metionina |
Na+ |
sodium ion |
NH3 |
ammonia |
NNP |
non-protein nitrogen |
pH |
hydrogen potential |
Phe |
phenylalanine |
PLP |
pyridoxal phosphate cofactor |
TAG |
triacylglycerols |
Thr |
threonine |
Trp |
tryptophan |
Val |
valine |
VLDL |
very low density lipoproteins |
β-HBA |
β- hydroxybutyrate |
The Rumen
The rumen is an anaerobic fermentation chamber (Armato et al., 2016), with an acid to neutral hydrogen potential (pH) of 5.5 to 7.0 (Jiang et al., 2017); this being the main determinant of the type and number of microorganisms (Resende Jr et al., 2019) and a temperature ranging from 38 to 42 ºC (Pourazad et al., 2016; Yazdi et al., 2016). The ruminal ecosystem is made up of three groups: I) bacteria, its concentration is 1 x 1010 and 1 x 1011/mL of ruminal fluid (Valente et al., 2016), and it is related to the energy content of the diet (Krause et al., 2013); Furthermore, non-protein nitrogen (NNP), like urea, must be converted to ammonia (NH 3) for it to be used by bacteria (DePeters y George, 2014; Wallace et al., 2017), transforming poor-quality protein into high quality protein (Puppel y Kuczyńska, 2016; Jin et al., 2018); group II) ciliated protozoa, its concentration ranges from 1 x 104 to 1 x 106/mL of rumen fluid, its function is to control the number of bacteria in the rumen (Francisco et al., 2019), they wrap starch that passes into the intestine, being a source of glucose (C 6 H 12 O 6) for the ruminant (Wallace et al., 2017), they do not synthesize protein from NNP (Jin et al., 2018) most are of the Isotricha or Entodinium genus (Gebreegziabher, 2016), and group III) fungi, they are found in a concentration of 1 x 103 to 1 x 105/mL of ruminal fluid, they have cellulolytic activity mainly in mature forages (Valente et al., 2016); some species are Neocallimastix frontalis, Caecomyces communis and Piromyces communis (Krause et al., 2013).
The Amilolytic-Cellulolytic Ruminal Microbiota and Anaerobic Fermentation
The degradation of polysaccharides present in forages is carried out by cellulolytic bacteria (Bacteriodes succinogenes, Ruminococcus albus), amilolytics (Bacteroides amylophylus, Streptococcus bovis), hemicellulolytics (Butyrivibrio fibrisolvens, Bacteroides ruminicolanos) and pectinolytics (Lachnospira multiparus, Succinivibrio dextrinosolvens (Valente et al., 2016), which obtain C6H12O6 and other monosaccharides such as xylose and fructose-6-phosphate, from cellulose and hemicellulose (Krause et al., 2013). The monomers are absorbed by microorganisms and they form a nicotinamide adenine dinucleotide in its reduced form (NADH+H +), pyruvate (C 3 H 3 O 3) and adenosine triphosphate (ATP) for its growth and maintenance (Wallace et al., 2017; Francisco et al., 2019). Fermentative digestion is anaerobic (Kittelmann et al., 2013; Yazdi et al., 2016), so C3H3O3 works as an electron collector, to generate NAD+ and ATP, removing NADH+H+ (Górka et al., 2017).
Volatile fatty acids (AGV): acetic (CH 3 -COOH), propionic (CH 3 -CH 2 -COOH) and butyric (CH 3 -CH 2 -CH 2 -COOH) are the main end products of fermentative digestion (Aydin et al., 2017; Li et al., 2019a); they are absorbed through the rumen wall and incorporated into the circulation through the portal vein (Resende Jr et al., 2019). They represent between 70-80% of the ruminant's energy fuel (Mikołajczyk et al., 2019).
The ruminal flora synthesizes CH3-COOH from the decarboxylation of C3H3O3 in acetyl coenzyme A, releasing a carbon (Gebreegziabher, 2016; Chishti et al., 2020). For the formation of CH3-CH2-CH2-COOH two acetyl coenzyme A are required (Górka et al., 2017; Resende Jr et al., 2019). There are two routes for the formation of CH3-CH2-COOH: I) direct reductive route, C3H3O3 passes to lactate, and this to acrylyl-coenzyme A A (Aydin et al., 2017), and II) random route, a carbon to C3H3O3 and the oxaloacetate formed is transformed into succinate; CH3-CH2-COOH is subsequently synthesized, losing one carbon and forming molecular dioxygen (Krehbiel, 2014; Gebreegziabher, 2016). In addition, carbon dioxide (CO 2) and methane (CH 4) are formed and are eliminated by belching (Teklebrhan et al., 2020; Toral et al., 2017). CH4 synthesis is necessary for the production of oxidized cofactors in the routes for the formation of CH3-COOH and CH3- CH2-CH2-COOH (Kozłowska et al., 2019). The bacteria responsible for this function are Methanobrevibacter ruminantium, Methanobacterium formicicum and Methanomicrobium mobile (Baruah et al., 2019).
Figure 1 shows AGV synthesis. The rumen concentration of CH3-COOH, CH3-CH2-COOH and CH3-CH2-CH2-COOH in animals fed on forage. It ranges 70: 20: 10% respectively, and in animals fed mainly with cereals it fluctuates 60: 30: 10% (Gebreegziabher, 2016).
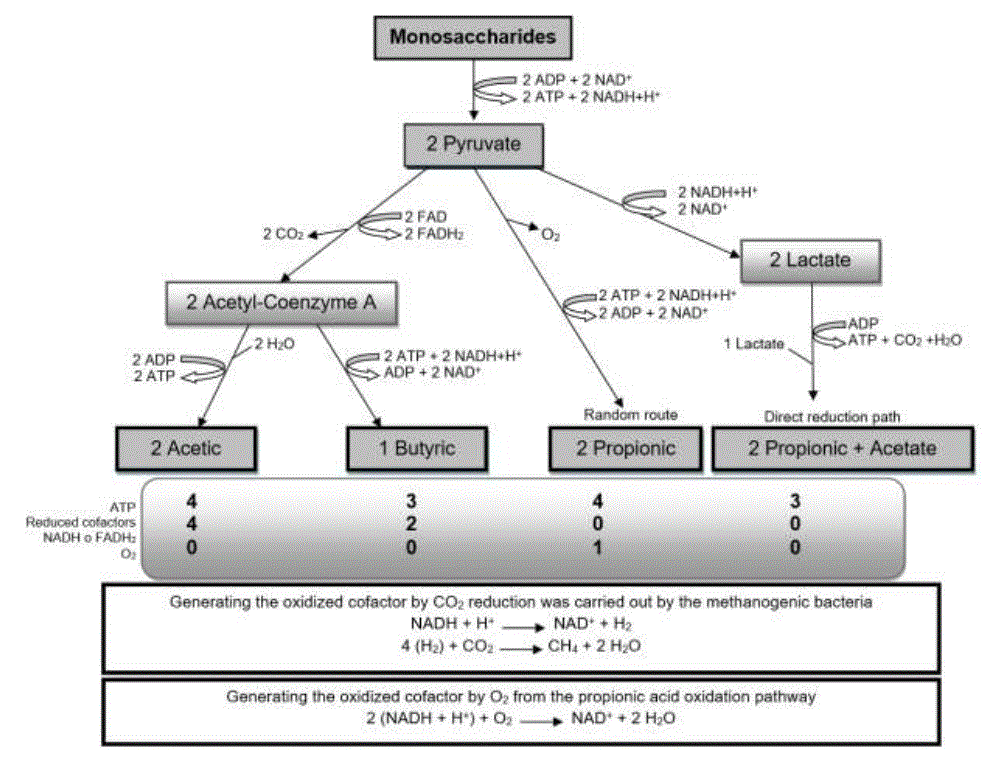
Source: synthesized information of (Gebreegziabher, 2016)
Figure 1 Synthesis of volatile fatty acids from monosaccharides in the rumen
The Proteolytic Ruminal Microbiota and Anaerobic Fermentation
The protein components supplied in the diet are fermented by proteolytic bacteria Bacteroides amylophylus, Bacteroides ruminicola, and some strains of Butyrivibriofibrisolvens (García et al., 2014), through their microbial proteases, releasing peptides (Alves et al., 2014; Rostom y Shine, 2018). These are absorbed by the microorganism, where the peptidases hydrolyze the peptide bonds, releasing aa, used to translate own proteins or catabolize them to release energy (Li et al., 2019b; Silva et al., 2016). The final product is NH3 (Khezri et al., 2016; Carvalho et al., 2019), which serves as a nitrogen substrate for bacteria (Valente et al., 2016). NH3 is absorbed by passive diffusion through potassium ion channels (K +), located in the rumen membrane (García et al., 2014), by portal circulation it reaches the liver where it is synthesized in urea (Rostom y Shine, 2018).
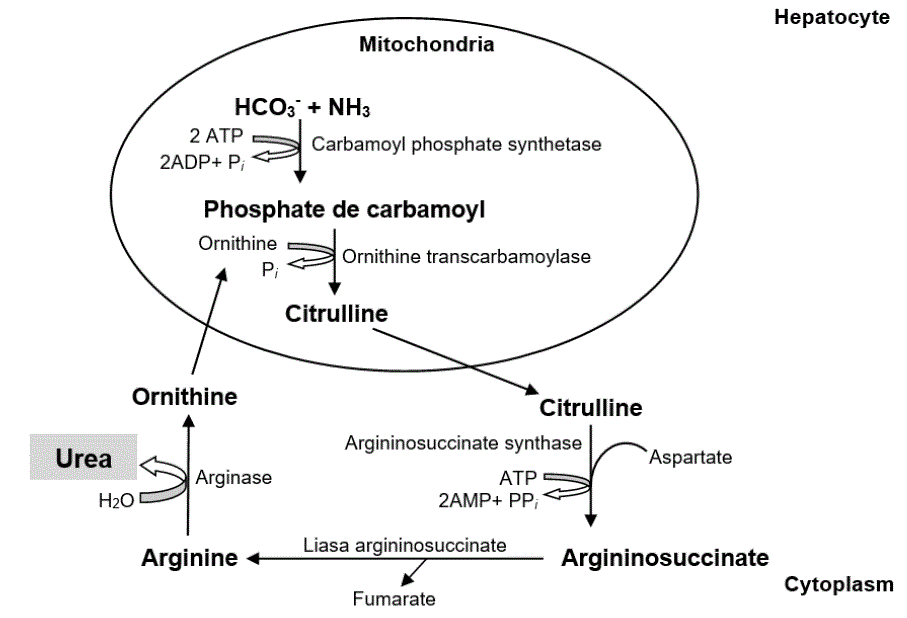
Urea synthesis begins in the mitochondrial matrix (Shi et al., 2019) with the binding of the hydrogen carbonate anion (HCO 3 -) and NH3, by means of carbamoyl phosphate synthetase. Carbamoyl phosphate binds to ornithine, via ornithine transcarbamoylase, generating citrulline. This is transported to the cytoplasm where it reacts with aspartate by means of argininosuccinate synthase, forming argininosuccinate. Subsequently, argininosuccinate lyase divides it, forming arginine (Arg) and fumarate (Hristov et al., 2019). Lastly, Arg catalyzes hydrolysis to synthesize ornithine, water (H 2 O) and urea (Gebreegziabher, 2016) (figure 2).
The urea goes back to the blood circulation where it has three metabolic routes: 1.) returns to the rumen via saliva or through the epitelial layers of rumen with the help of transport protein UT-B to be converted in NH3 (García et al., 2014; Carvalho et al., 2019), 2) excreted in the urine or feces (Schuba et al., 2017; Li et al., 2019b) or, 3) to be part of NNP of milk (Alves et al., 2014; Jin et al., 2018) (figure 3).
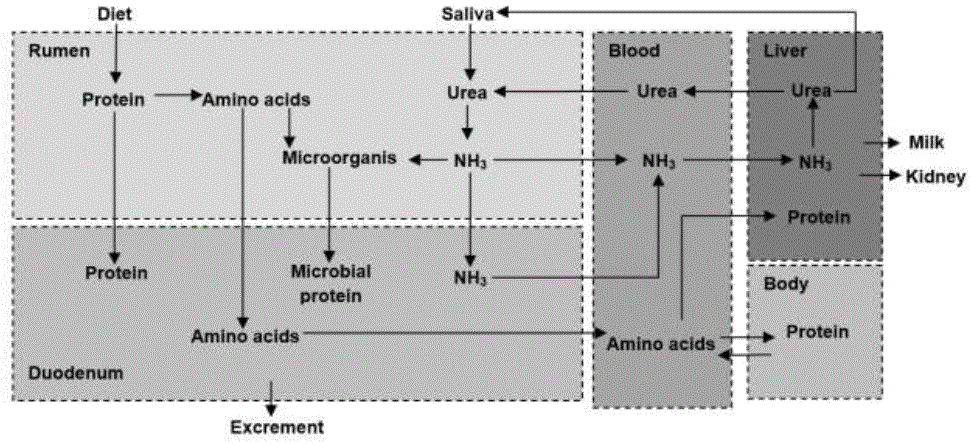
Source: synthesized information of (Li et al., 2019b)
Figure 3 General metabolism of proteins in the rumian
The Lipolytic Ruminal Microbiota and Anaerobic Fermentation
The microorganisms in charge of catabolizing the lipid components of the diet are: Anaerovibrio lipolytica, Butyrivibrio fibrisolvens, Treponema bryantii, Eubacterium spp., Fusocillus spp. and Micrococcus spp. (Valente et al., 2016). Bacterial lipases by hydrolysis release unesterified fatty acids (AGNE) and glycerol (Prieto et al., 2016); In addition, amino alcohols (derived from phospholipids) and galactose (from galactolipids) (Toral et al., 2018). Glycerol, amino alcohols and galactose are metabolized to AGV (Silva et al., 2014; van Cleef et al., 2018). The AGNE that are free in the rumen, carry out a microbial hydrogenation process (Tran et al., 2017; Toral et al., 2017), result of the addition of hydrogen to saturated fatty acids, to form unsaturated fatty acids with double bonds (Francisco et al., 2019). This mechanism is another way to eliminate the hydrogens that result from the catabolism of the polysaccharides (Osorio et al., 2015; Prieto et al., 2016).
The absorption of AGV is carried out in the rumen wall (80%), in omasum (10%), and the rest passes to the abomasum to be absorbed in the duodenum (Yazdi et al., 2016). AGVs passively diffuse into the ruminal epithelium (Agarwal et al., 2015; Yohe et al., 2019). The hydrogen necessary for the AGVs to dissociate in the epithelium is donated by carbon dioxide (H 2 CO 3), forming CO2 and H2O, from the dissociation a hydrogen is obtained to bind to the AGVs and a HCO3- molecule is formed in the lumen of the rumen. Therefore, this process helps buffer the rumen pH (Wang et al., 2016).
The absorption of AGV is carried out in the same way for all, although inside the epithelial cells of the rumen its conformation changes (Qumar et al., 2016). A part of the CH3-COOH is completely oxidized inside the cells, as an energy source; while the rest is absorbed without being altered, passing to the liver through the portal vein (Loncke et al., 2015). 80% of the CH3-COOH that reaches the liver escapes oxidation, passing into the general circulation to be used by other tissues (Qumar et al., 2016).
In the cytoplasm, the conversion of CH3-COOH to acetyl-Coenzyme A is catalyzed by acetyl-Coenzyme A synthetase (Chishti et al., 2020). Most of it is oxidized in the Krebs cycle or is used for fatty acid synthesis in hepatocytes (Yohe et al., 2019). A fraction of CH3-CH2-COOH is degraded and converted to lactate (2-5%) before or during absorption; the rest passes in the portal circulation to the liver, where the hepatocytes synthesize it in C6H12O6, via glycogenesis (Loncke et al., 2015). To enter the Krebs cycle, propionyl- Coenzyme A through propionyl-Coenzyme A carboxylase, forms methylmalonyl- Coenzyme A, and then succinyl-Coenzyme A is formed (Gebreegziabher, 2016). CH3-CH2-CH2-COOH is converted almost entirely to β-hydroxybutyrate (β-HBA) in the rumen mucosa (Agarwal et al., 2015). This ketone body represents 80% of the ketones formed (Górka et al., 2017). CH3-COOH and β-HBA are used for the synthesis of fatty acids in adipose tissue and the mammary gland (García et al., 2015; Song et al., 2018).
Postruminal Digestion and Absorption
Although the ruminant is characterized by microbial fermentation in the rumen (Hristov et al., 2019), post-ruminal digestion is vital, since it has lipids, proteins and some non- structural polysaccharides that escape from fermentation (Agarwal et al., 2015) The unfermented food along with microbial protein, passes to the omasum through the reticulo-omasal hole, where AGV, NH3, H2O, sodium ion (Na +) and K+ are absorbed (Hussain et al., 2013; Freitas Jr et al., 2019). Subsequently, they pass to the abomasum containing hydrochloric acid (HCl) and pepsin (Rotta et al., 2014). Food is mixed, passing into the duodenum (Hristov et al., 2019). The starch and disaccharides that escape from the ruminal digestion are hydrolyzed by pancreatic amylases, obtaining monosaccharides (Rotta et al., 2014).
Absorption takes place in the villi of the enterocytes (Harmon, 2009). Monosaccharides are transported against their concentration gradient by means of the Na+ co-transporter (Harmon y Swanson, 2020). The ATPase-Na+- K+ pump creates the energy-contributing Na+ concentration gradient (Bergman et al., 2019).
Another form of transport for C6H12O6 is the GLUT2 transporter (Harmon, 2009). The protein that reaches the small intestine comes from the diet that escapes from fermentation, endogenous protein (García et al., 2015) and that contained in the microorganisms that are linked to food (Batista et al., 2016; Golshan et al., 2019). Catabolism begins in the abomasum due to pepsin and acid hydrolysis; later in the duodenum by pancreatic and duodenal enzymes (trypsinase, chymotrypsinase and carboxypeptidase), which break peptide bonds to release aa and small peptides for their absorption in jejunum and ileum (Emery, 2015; Hristov et al., 2019). Absorption consists of transport through Na+ dependent, energy consumption is associated with the continuous flow of Na+ to the outside, as a result of the activity of the ATPase-Na+-K+ pump (Silva et al., 2016).The Na+ that enters the cell in favor of a concentration gradient, is bound to an aa molecule through the cell membrane (Emery, 2012; Rostom y Shine, 2018).
The lipids that reach the abomasum in the form of AGNE represent between 70 and 80%, the rest are phospholipids of microbial origin (Aibibula et al., 2015; Toral et al., 2018). The latter are emulsified by bile salts and hydrolyzed by pancreatic lipases to release AGNE (Dawson y Karpen, 2015; Kohan et al., 2015). The micelle is formed from bile salts, saturated AGNE, triacylglycerols (TAG) and lecithin (Cao et al., 2018), transporting itself to the villi of the enterocytes (Park et al., 2019). AGNE of less than 12 carbons are absorbed and transported by portal vein to the liver linked by non-covalent bonds in albumin (ALB) (Dawson y Karpen, 2015). In contrast, AGNE of 12 or more carbons are esterified to form TAGs and phospholipids (Vargas, 2019). TAGs, small amounts of mono and diacylglycerols, phospholipids and cholesterol (COL) are bound to apoproteins to form chylomicrons and very low density lipoproteins ( VLDL ), which leave the lymphatic system, to be incorporated into the bloodstream (Kohan et al., 2015; Prieto et al., 2016). Lipids are absorbed by diffusion or pinocytosis (Walther y Farese Jr, 2012).
Monosaccharide Metabolism in Ruminants
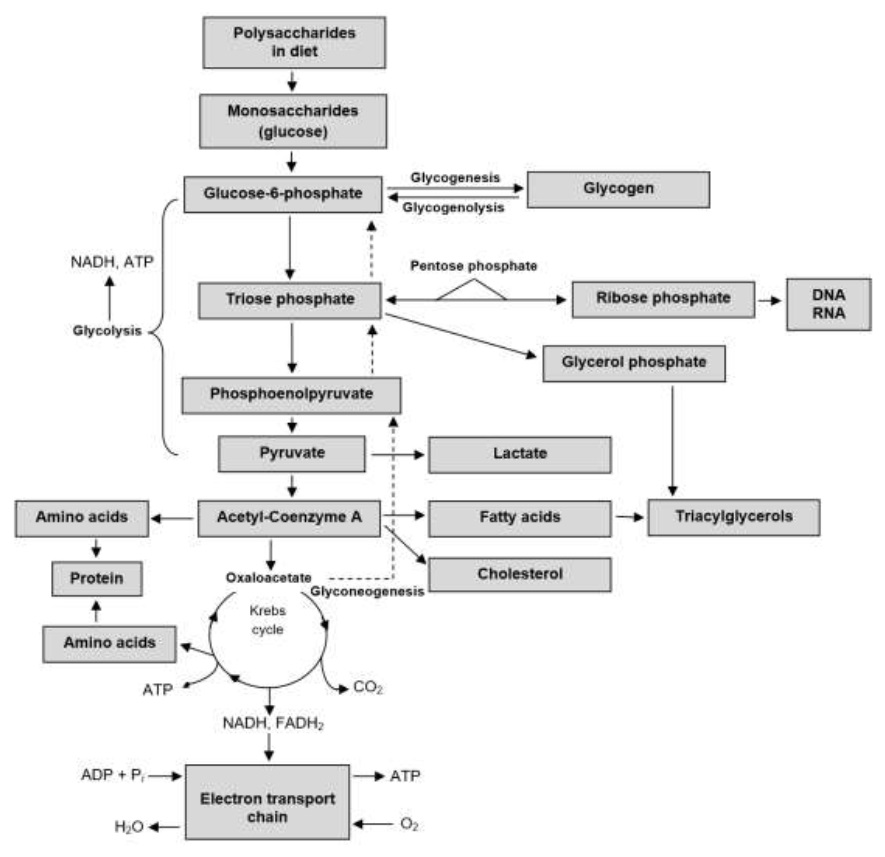
The blood stream is the means by which the absorbed nutrients are directed to the liver and other organs for catabolism or anabolism, depending on cellular need (Goyal y Longo, 2015). Enzymes play a very important role in metabolism, as they are catalytic proteins for specific reactions (Jindal y Warshel 2017); Without them, biological reactions would be very slow for cell life (Ramsay et al., 2019). Its function is to temporarily bind to a molecule, to apply atomic changes (Menger y Nome, 2019). Monosaccharide metabolism revolves around the supply and destination of C6H12O6, with this monomer being the main source of energy for cells (Hooijberg et al., 2017). The catabolic route of C6H12O6 is glycolysis, carried out in the cellular cytoplasm (Dashty, 2013) This process consists of eight reactions: 1) glucose (C6H12O6) enters the cytoplasm to be phosphorylated (addition of a phosphate group), starting from ATP. This reaction is catalyzed by hexokinase. The resulting glucose-6-phosphate (C6H11O9P) (aldohexose) abounds in all cells, since the vast majority of C6H12O6 that enters the cytoplasm ends up being phosphorylated, in order to prevent that it can cross the cytoplasmic membrane back and diffuse into the extracellular medium (Donnelly y Finlay, 2015); 2) C6H11O9P has isomerization [one molecule is transformed into another that has the same atoms, but arranged differently - the carbonyl group (C=O) - is replaced] and is transformed into fructose-6-phosphate (ketohexose) . Glucose-6-phosphate isomerase catalyzed reaction (Dashty, 2013); 3) fructose-6-phosphate, is phosphorylated from ATP, at carbons 1 and 6 to give fructose- 1,6-bisphosphate. Phosphofructokinase catalyzed reaction (Ashrafi y Ryan, 2017)Ñ 4) Fructose-1,6-bisphosphate is divided into two: glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. Aldose catalyzed reaction (Watts y Ristow, 2017); 5) triose phosphate isomerase catalyzes the conversion of dihydroxyacetone phosphate to obtain more glyceraldehyde-3-phosphate (Bommer et al., 2020); 6) glyceraldehyde-3-phosphate is oxidized and phosphorylated, at carbons 1 and 6 forming 1,3-bisphosphoglycerate by glyceraldehyde-phosphate dehydrogenase (Poher et al., 2018). Subsequently, it transfers its phosphate group, to synthesize ATP and it is transformed into 3-phosphoglycerate. Phosphoglycerate kinase catalyzed reaction (Dashty, 2013); 7) 3-phosphoglycerate exhibits isomerization of C3 to C2 and it is transformed into 2-phosphoglycerate by phosphoglycerate mutase (Donnelly y Finlay, 2015). Subsequently, enolase promotes the formation of a double bond, eliminating an H2O molecule and forming phosphoenolpyruvate (Bommer et al., 2020) and 8) phosphoenolpyruvate transfers its phosphate group, to synthesize ATP and it is transformed into C3H3O3, a reaction catalyzed by pyruvate kinase (figure 4).
C3H3O3 leaves the cytoplasm and enters the mitochondrial matrix, using the proton-motor force generated by the respiratory chain (Poher et al., 2018). For each C6H12O6, two C3H3O3, two ATP, two NADH+H+, two hydrogenions and two H2O molecules are generated (Dashty, 2013; Watts y Ristow, 2017). Aerobic cells metabolize C3H3O3 to acetyl-Coenzyme A, by means of pyruvate dehydrogenase (Edinburgh et al., 2017), allowing its entry into the Krebs cycle for its participation in oxidative phosphorylation (Bergman et al., 2019).
For each acetyl-Coenzyme A that enters the Krebs cycle, 12 ATP are produced. This process is an essential source of intermediaries for other metabolic pathways, eg. eg, glycogenogenesis in the liver and striated muscle (Dashty, 2013; Edinburgh et al., 2017), the pentose phosphate pathway (Figure 4) and lipid synthesis and aa. The pentose phosphate pathway, is an alternate metabolic pathway that does not produce ATP (Kohan et al., 2015), synthesizes reducing equivalents such as nicotinamide adenine dinucleotide (NADPH), for the de novo synthesis of fatty acids, steroids, maintenance of glutathione for antioxidant activity (Chen et al., 2016) and ribose sources for the synthesis of nucleic acids and nucleotides (Norris et al., 2016).
The triose phosphate intermediate of glycolysis forms the glycerol moiety in TAGs (Edinburgh et al., 2017). On the other hand, C3H3O3 and Krebs cycle intermediaries supply the carbon skeletons for the synthesis of aa (Valdebenito et al., 2016) and acetyl-Coenzyme A is the precursor of AGNE, COL and steroid hormones (Edinburgh et al., 2017). Gluconeogenesis synthesizes C6H12O6 from lactate, aa and glycerol (Cantalapiedra et al., 2015; Campos et al., 2018), in the cytoplasm and mitochondria of hepatocytes (Chen et al., 2016; Qaid y Abdelrahman, 2016). In this route, six ATP are consumed for each C6H12O6 produced (Gebreegziabher, 2016) and the CH3-CH2-COOH propionate is the only glycogenic AGV (Wallace et al., 2017).
The importance of glycogenesis in ruminants (figure 4), is due to the fact that small amounts of C6H12O6 are absorbed by the body from the digestive tract and its ability to store glycogen in the liver is limited (Qaid y Abdelrahman, 2016).
Fatty Acid Metabolism in Ruminants
Lipid metabolism mainly depends on fatty acids and COL (Watts y Ristow, 2017). The source of long-chain AGNE is provided by diet or by de novo synthesis from acetyl- Coenzyme A, which is derived from monosaccharides or aa carbon skeletons (Walther y Farese Jr, 2012). The synthesis of fatty acids begins in the mitochondria with the formation of acetyl-Coenzyme A, from the oxidation of CH3-COOH and CH3-CH2-CH2-COOH (Vargas, 2019). Within the mitochondria, acetyl-Coenzyme A is produced; however, the mitochondrial membrane is impervious to its passage. Therefore, the tricarboxylate system and the action of citrate synthetase are required to convert acetyl-Coenzyme A to citrate and allow its passage into the cell cytoplasm (Civeira et al., 2013; Nunes-Nesi et al., 2013).
Once in the cytoplasm, the citrate is transformed again into acetyl-Coenzyme A by means of ATP-citrate lyase, also obtaining oxaloacetate and adenosine diphosphate (ADP) (Walther y Farese Jr, 2012). As the process for the synthesis of fatty acids is endergonic (it accumulates energy from carbons), acetyl-Coenzyme A presents carboxylation [a carboxyl group (COOH) is structured in the molecule], through its union with HCO3 - in a reaction catalyzed by acetyl-Coenzyme A carboxylase (García et al., 2014).
Oxaloacetate is reduced by malate dehydrogenase to malate, and this in turn is converted to C3H3O3 by malate dehydrogenase, giving the electron donor nicotinamide adenine dinucleotide phosphate in its reduced form (NADPH+H + ) (Watts y Ristow, 2017; Vargas, 2019). From malonyl-Coenzyme A, the synthesis of fatty acids is carried out by elongation, using fatty acid synthase (Du et al., 2018). This protein complex performs synthesis, reduction, dehydration, and reduction again, condensing the malonyl-Coenzyme A groups with acetyl-Coenzyme A (Civeira et al., 2013; Norris et al., 2016). In the elongation, groups of two carbons are added to the fatty acid, obtaining palmitic (C16:0) as the final fatty acid (Shi et al., 2018)..
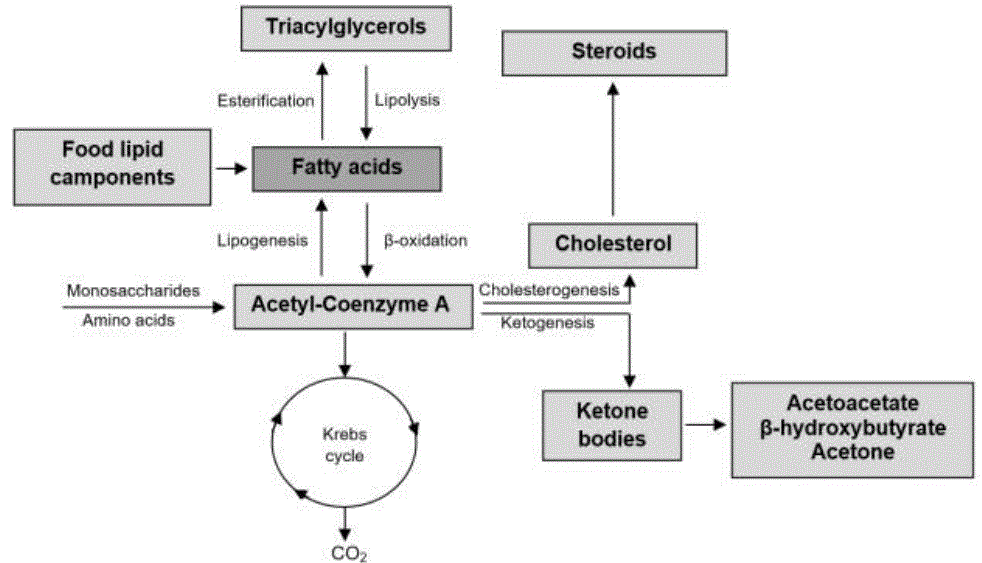
Fatty acids (figure 5) can be oxidized to acetyl-Coenzyme A by mitochondrial β-oxidation, or esterified with glycerol to form TAG and function as the body's main energy reserve (Osorio et al., 2015). TAG synthesis begins with the formation of glycerol-3-phosphate (Fong et al., 2016), later acyl-Coenzyme A fatty synthase activates fatty acids and three of them are esterified to the molecule (Civeira et al., 2013).
In TAG catabolism, the ester bonds at C1 or at C3 are hydrolyzed, obtaining AGNE. Hormone sensitive lipase catalyzed reaction (McFadden, 2020). AGNE are transported in the bloodstream, through non-covalent binding with ALB, where they are captured and oxidized by myocytes or hepatocytes, or stored by adipocytes (Edinburgh et al., 2017). The β-oxidation is carried out in the mitochondrial matrix (Morita et al, 2016), being carried out by means of the activation of fatty acids by means of thiosinase in acyl-Coenzyme A (Walther y Farese Jr, 2012); this process requires ATP to form adenylyl (Fukao et al., 2014). Activated acyl-Coenzyme A enters the mitochondrial matrix through carnitine palmitoyltransferase (Nunes-Nesi et al., 2013; Morita et al, 2016), andi t is oxidized by fatty acyl-Coenzyme A dehydrogenase (Houten y Wanders, 2010). Hydrogen atoms are accepted by flavin-adenine dinucleotide (FAD) which is reduced to FADH2 (Norris et al., 2016). Subsequently, enoyl-Coenzyme A hydratase introduces H2O into the newly formed double bond between C2 and C3 (Kong et al., 2017) and β-hydroxyacyl Coenzyme A dehydrogenase forms 3-ketoacyl-Coenzyme A (Walther y Farese Jr, 2012; Martines et al., 2017). The two removed atoms are transferred to NAD+ generating NADH+H+ (Kohan et al., 2015).
Finally thiolase divides C1 and C2 from 3-ketoacyl-Coenzyme A, releasing acetyl- Coenzyme A (Martines et al., 2017), this shortens the two-carbon acyl-Coenzyme A chain, requiring another Coenzyme A, to finish the newly shortened molecule (Kong et al., 2017). These steps are repeated until leaving a four-carbon acyl-Coenzyme A, where the four steps are repeated, only that instead of releasing one acetyl-Coenzyme A two are released (Civeira et al., 2013).
When it comes to an odd fatty acid the penultimate repeat leaves a five-carbon fatty acyl- Coenzyme A and it undergoes the previous four steps, but the final two steps give one molecule of acetyl-Coenzyme A and one molecule of propionyl- Three carbon coenzyme A (Houten y Wanders, 2010). Acetyl-Coenzyme A as a product of the β-oxidation of fatty acids, can have three destinations: a) enter the Krebs cycle to oxidize to CO2 and H2O for energy release (Fukao et al., 2014; Panov et al., 2014); b) serve as a precursor for the synthesis of COL and other steroids (Walther y Farese Jr, 2012), and c) participate in ketogenesis (Watts y Ristow, 2017). The ketone bodies acetoacetate (AcAc), β-HBA and acetone (Garzón y Espinosa, 2018), serve as a substrate for the production of ATP (McFadden, 2020). They are synthesized in the liver, in low concentrations, but when intracellular C6H12O6 decreases, their synthesis rises (Norris et al., 2016).
Ketogenesis takes place in the mitochondrial matrix (Fukao et al., 2014). When hepatic glycogen reserves decrease, the activity of carnitine palmitoyltransferase is stimulated, causing the transport of AGNE into the hepatic mitochondria (Walther y Farese Jr, 2012), where a series of successive β-oxidations is carried out, leading to the formation of acetyl- Coenzyme A (McFadden, 2020). This molecule is combined with oxaloacetate for its entry into the Krebs cycle (García et al., 2015). If this oxidation is complete, CO2 and hydrogen atoms will be released, which will donate their electrons to carry out oxide reduction reactions, which will culminate in the formation of H2O and ATP (McFadden, 2020).
If oxaloacetate is reduced by acetyl-Coenzyme A, it accumulates within the hepatic mitochondria (Walther y Farese Jr, 2012); reason why two acetyl-Coenzyme A molecules react to form acetoacetyl-Coenzyme A, catalyzed by thiolase (Fukao et al., 2014). Acetoacetyl-Coenzyme A binds with another acetyl-Coenzyme A molecule to form β- hydroxy-β-methylglutaryl-CoA, catalyzed by 3-hydroxy-3-methylglutaryl-CoA synthase (Norris et al., 2016). Finally, the molecule is metabolized in AcAc (figure 5) and leaves the mitochondria to the cytoplasm, where it can be reduced in β-HBA or decarboxylated, up to acetone (García et al., 2015).
Amino Acid Metabolism in Ruminants
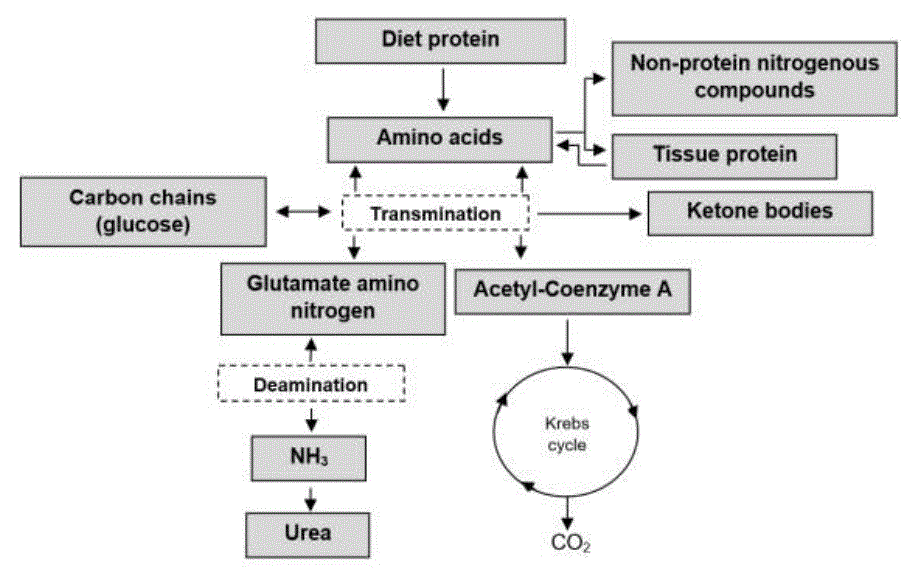
The metabolism of aa involves transamination and deamination (Dong et al., 2016), necessary reactions for the anabolism and catabolism of proteins (Golshan et al., 2019). The aa Arg, histidine (His), isoleucine (Ile), leucine (Leu), lysine (Lys), methionine (Met), phenylalanine (Phe), threonine (Thr), tryptophan (Trp) and valine (Val), are mostly produced by ruminal fermentation (Zhou et al., 2019). The aa are composed of an amino group (-NH 2) and a COOH group; in addition to an R side chain, which gives them hydrophilic, hydrophobic, acidic, basic and aromatic properties (Rostom y Shine, 2018). Transamination is carried out by aminotransferases, the -NH2 group is transferred from an acidic aa to a ketoacid aa (Zhou et al., 2019; Batista et al., 2016). Aminotransferases are located in the cytoplasm and mitochondria, having two types of specificity: I) the type of aa that donates -NH2 (Emery, 2015) and II) the keto acid that accepts -NH2 (Dong et al., 2016). Although enzymes vary depending on the type of aa they bind, most use glutamic (Glu) as a -NH2 donor (Rostom y Shine, 2018).
These reactions require the pyridoxal phosphate cofactor (PLP) (Witus et al., 2013). In oxidative deamination the aa lose the -NH2, a reaction catalyzed by glutamate dehydrogenase (Dong et al., 2016). The resulting carbon skeletons are degraded to one of seven possible metabolic products: acetyl-Coenzyme A, acetoacetyl-Coenzyme A, C3H3O3, ketoglutarate, succinyl-Coenzyme A, fumarate, or oxaloacetate (Rostom y Shine, 2018). The aa's that degrade from acetyl-Coenzyme A to acetoacetyl-Coenzyme A are known as ketogens (Lys and Leu) (Batista et al., 2016). The carbon skeletons of glycogenic aa degrade to C3H3O3 or a Krebs cycle intermediate, but can also be converted to C6H12O6 by glycogenesis (Emery, 2012). The NH3 resulting from the deamination of the aa (figure 6) is transported to the periportal hepatocytes to participate in ureogenesis (García et al., 2014).
CONCLUSION
The scientific elements presented on anabolism and catabolism of nutrients show that intestinal absorption of glucose in ruminants is limited. Therefore, the ruminal microbiota plays an important role in the transformation, assimilation, and synthesis of each of the biochemical monomers; elements of vital importance in glycogenesis, proteogenesis, ureogenesis, lipogenesis and ketogenesis; metabolic processes that confer information on alterations and homeostatic adjustments in ruminants.
LITERATURA CITADA
Agarwal U, Hu Q, Baldwin RL, Bequette BJ. 2015. Role of rumen butyrate in regulation of nitrogen utilization and urea nitrogen kinetics in growing sheep. Journal of Dairy Science. 93(1):2382-2390. ISSN: 0022-0302. https://doi.org/10.2527/jas.2014-8738 [ Links ]
Aibibula Y, Halidai R, Masaaki H, Meiji O. 2015. Rumen degradability and post-ruminal degestion of nitrogen and amino acids by cows grazing temperate pasture. Asian Agricultural Research. 7(5):72-78. ISSN: 1011-2367. http://dx.doi.org/10.22004/ag.econ.207047 [ Links ]
Alves EM, Magalhães DR, Freitas MA, Dos Santos EJ, Pereira MLA, Pedreira MS. 2014. Nitrogen metabolism and microbial synthesis in sheep fed diets containing slow release urea to replace the conventional urea. Acta Scientiarum: Animal Sciences. 36(1):55-62. ISSN: 1807-8672. https://doi.org/10.4025/actascianimsci.v36i1.21377 [ Links ]
Armato LM, Gianesella M, Morgante M, Fiore E, Rizzo M, Giudice E, Piccione G. 2016. Rumen volatile fatty acids x dietary supplementation with live yeast and yeast cell wall in feedlot beef cattle. Acta Agriculturae Scandinavica: Animal Science. 66(2):119-124. ISSN: 0906-4702 http://dx.doi.org/10.1080/09064702.2016.1272628 [ Links ]
Ashrafi G, Ryan TA. 2017. Glucose metabolism in nerve terminals. Current Opinion in Neurobiology. 45(1):156-161. ISSN: 0959-4388. http://dx.doi.org/10.1016/j.conb.2017.03.007 [ Links ]
Aydin S, Yıldırım E, Ince O, Ince B. 2017. Rumen anaerobic fungi create new opportunities for enhanced methane production from microalgae biomass. Algal Research. 23(1):150-160. ISSN: 2211-9264. http://dx.doi.org/10.1016/j.algal.2016.12.016 [ Links ]
Baruah L, Malik PK, Kolte AP, Goyal P, Dhali A, Bhatta R. 2019. Rumen methane amelioration in sheep using two selected tanniferous phyto-leaves. Carbon Management. 10(3):299-308. ISSN: 1758-3004. https://doi.org/10.1080/17583004.2019.1605480 [ Links ]
Batista ED, Detmann E, Titgemeyer EC, Valadares-Filho SC, Valadares RFD, Prates LL, Rennó LN, Paulino MF. 2016. Effects of varying ruminally undegradable protein supplementation on forage digestion, nitrogen metabolism, and urea kinetics in Nellore cattle fed low-quality tropical forage. Journal Animal Science. 94(1):201-216. ISSN: 1525-3163. https://doi.org/10.2527/jas.2015-9493 [ Links ]
Bergman RN, Piccinini F, Kabir M, Ader M. 2019. Novel aspects of the role of the liver in carbohydrate metabolism. Metabolism Clinical and Experimental. 99(1):119-125. ISSN: 0026-0495. https://doi.org/10.1016/j.metabol.2019.05.011 [ Links ]
Bommer GT, Schaftingen EV, Veiga-da-Cunha M. 2020. Metabolite repair enzymes control metabolic damage in glycolysis. Trends in Biochemical Sciences. 45(3):16-32. ISSN: 0968-0004. https://doi.org/10.1016/j.tibs.2019.07.004 [ Links ]
Campos GR, Correa-Orozco A, Zambrano BGL, Ospina CA. 2018. Alteraciones bioquímicas y metabólicas en el período de transición en vacas lecheras. Revista de Investigación Agraria y Ambiental. 9(2):166-179. ISSN: 2145-6097.https://doi.org/10.22490/21456453.2123 [ Links ]
Cantalapiedra-Hijar G, Ortigues-Marty I, Sepchat B, Agabriel J, Huneau JF, Fouillet H. 2015. Diet-animal fractionation of nitrogen stable isotopes reflects the efficiency of nitrogen assimilation in ruminants. British Journal of Nutrition. 113(1):1158-1169. ISSN: 0007-1145. https://doi.org/10.1017/S0007114514004449 [ Links ]
Cao YC, Yang XJ, Guo L, Zheng C, Wang DD, Cai CJ, Yao JH. 2018. Regulation of pancreas development and enzymatic gene expression by duodenal infusion of leucine and phenylalanine in dairy goats. Livestock Science. 216(1):9-15. ISSN: 1871-1413. https://doi.org/10.1016/j.livsci.2018.03.010 [ Links ]
Carvalho IPC, Doelman J, Martín-Tereso J.2019. Post-ruminal non-protein nitrogen supplementation as a strategy to improve fibre digestion and N efficiency in the ruminant. Journa of Animal Physiology Animal Nutrition. 104(1):64-75. ISSN: 1439-0396. https://doi.org/10.1111/jpn.13233 [ Links ]
Civeira F, Baila-Rueda L, Castro-Orós I, Mateo-Gallego R, Cenarro A.2013. Novedades en el metabolismo lipídico. Revista Nefroligía. 4(4):9-17. ISSN: 0211-6995. http://dx.doi.org/10.3265/NefrologíaSuplementoExtraordinario.pre2013.Nov.12338 [ Links ]
Chen L, Tuo B, Dong H. 2016. Regulation of intestinal glucose absorption by ion channels and transporters. Nutrients. 8(43):2-13. ISSN: 2072-6643. https://doi.org/10.3390/nu8010043 [ Links ]
Chishti GA, Salfer IJ, Suarez-Mena FX, Harvatine KJ, Heinrichs AJ. 2020. Short communication: Relationships between physical form of oats in starter, rumen pH, and volatile fatty acids on hepatic expression of genes involved in metabolism and inflammation in dairy calves. Journal of Dairy Science. 103(1):10-18. ISSN: 0022-0302. https://doi.org/10.3168/jds.2019-16296 [ Links ]
Dashty M. 2013. A quick look at biochemistry: Carbohydrate metabolism. Clinical Biochemistry. 46(1):1339-1352. ISSN: 0009-9120. http://dx.doi.org/10.1016/j.clinbiochem.2013.04.027 [ Links ]
Dawson PA, Karpen SJ. 2015. Intestinal transport and metabolism of bile acids. Journal of Lipid Research. 56(1):1085-1099. ISSN: 0022-2275. https://doi.org/10.1194/jlr.r054114 [ Links ]
DePeters EJ, George LW. 2014. Rumen transfaunation. Immunology Letters. 162(1):69-76. ISSN: 0165-2478. http://dx.doi.org/10.1016/j.imlet.2014.05.009 [ Links ]
Dong J, Jeong HJ, Ueda H. 2016. Preparation of quenchbodies by protein transamination reaction. Journal of Bioscience and Bioengineering. 122(1):125-130. ISSN: 1389-1723. http://dx.doi.org/10.1016/j.jbiosc.2015.12.010 [ Links ]
Donnelly RP, Finlay DK. 2015. Glucose, glycolysis and lymphocyte responses. Molecular Immunology. 68(1):513-519. ISSN: 0161-5890. http://dx.doi.org/10.1016/j.molimm.2015.07.034 [ Links ]
Du X, She T, Wang H, Qin X, Xing D, Ye Q, Shi Z, Fang Z, Zhu Y, Yang Y, Peng Z, Zhao C, Lv B, Li X, Liu G, Li X. 2018. Adaptations of hepatic lipid metabolism and mitocondria in dairy cows with mild fatty liver. Journal Dairy Science. 101(10):9544-9558. ISSN: 0022-0302. https://doi.org/10.3168/jds.2018-14546 [ Links ]
Edinburgh RM, Betts JA, Burns SF, González TJ. 2017. Concordant and divergent strategies to improve postprandial glucose and lipid metabolism. Nutrition Bulletin. 42(1):113-122. ISSN: 1467-3010. https://doi.org/10.1111/nbu.12259 [ Links ]
Emery PW. 2012. Basic metabolism: protein. Surgery. 30(5):209-213. ISSN: 0039-6060. https://doi.org/10.1016/j.mpsur.2012.02.008 [ Links ]
Emery PW. 2015. Basic metabolism: protein. Surgery. 33(4):143-147. ISSN: 0039-6060. https://doi.org/10.1016/j.mpsur.2015.01.008 [ Links ]
Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. 2016. GPIHBP1 and plasma triglyceride metabolism. Trends in Endocrinology & Metabolism. 27(7):445-469. ISSN: 1043-2760. http://dx.doi.org/10.1016/j.tem.2016.04.013 [ Links ]
Francisco AE, Santos-Silva JM, Portugal APV, Alves SP, Bessa RJB. 2019. Relationship between rumen ciliate protozoa and biohydrogenation fatty acid profile in rumen and meat of lambs. PLoS ONE. 14(9):221-243. ISSN: 1932-6203. https://doi.org/10.1371/journal.pone.0221996 [ Links ]
Freitas Jr JE, Bettero VP, Zanferari F, Del Valle TA, De Paiva PG, De Jesus EF, Takiya CS, Leite LC, Dias M, Rennó FP. 2019. Ruminal fatty acid outflow in dry cows fed different sources of linoleic acid: reticulum and omasum as alternative sampling sites to abomasum. Archives of Animal Nutrition. 70(3):171-193. ISSN: 1745-039X. https://doi.org/10.1080/1745039X.2019.1595886 [ Links ]
Fukao T, Mitchell G, Sass JO, Hori T, Orii K, Aoyama Y. 2014. Ketone body metabolism and its defects. Journal of Inherited Metabolic Disease. 37(1):541-551. ISSN: 0141-8955. http://dx.doi.org/10.1007/s10545-014-9704-9 [ Links ]
García CAC, Montiel RLA, Borderas TF, Girard V. 2015. Relationship between β- hydroxybutyrate and the fat: protein ratio of milk during early lactation in dairy cows. Archivos de Medicina Veterinaria. 47(1):21-25. ISSN: 0301-732X. http://dx.doi.org/10.4067/S0301-732X2015000100005 [ Links ]
García CAC, Montiel RLA, Borderas TF. 2014. Grasa y proteína de la leche de vaca: componentes, síntesis y modificación. Archivos de Zootecnia. 63(1):85-105. ISSN: 1885-4494. https://doi.org/10.21071/az.v63i241.592 [ Links ]
Garzón AAM, Espinosa OJ. 2018. Epidemiología de la cetosis en bovinos: una revisión. Revista CES Medicina Veterinaria y Zootecnia. 13(1):42-61. ISSN: 1900-9607. http://dx.doi.org/10.21615/cesmvz.13.1.4 [ Links ]
Gebreegziabher Z. 2016. Factors affecting feed intake and its regulation mechanisms in ruminants. A Review. International Journal of Livestock Research. 6(4):19-40. ISSN: 2277-1964. https://doi.org/10.5455/ijlr.20160328085909 [ Links ]
Ginane C, Bonnet M, Baumont R, Revell DK. 2015. Feeding behaviour in ruminants: a consequence of interactions between a reward system and the regulation of metabolic homeostasis. Animal Production Science. 55(1):247-260. ISSN: 1836-0939. http://dx.doi.org/10.1071/AN14481 [ Links ]
Golshan S, Pirmohammadi R, Khalilvandi-Behroozyar H. 2019. Microwave irradiation of whole soybeans in ruminant nutrition: Protein and carbohydrate metabolism in vitro and in situ. Veterinary Research Forum. 10(4):343-350. ISSN: 2008-8140. https://dx.doi.org/10.30466%2Fvrf.2019.35896 [ Links ]
Górka P, Śliwiński B, Flaga J, Wieczorek J, Godlewski MM, Wierzchoś E, Zabielski R, Kowalski ZM. 2017. Effect of butyrate infusion into the rumen on butyrate flow to the duodenum, selected gene expression in the duodenum epithelium, and nutrient digestion in sheep. Journal Animal Science. 95(1):2144-2155. ISSN: 1525-3163. https://doi.org/10.2527/jas.2016.1218 [ Links ]
Goyal R, Longo LD. 2015. Metabolic profiles in ovine carotid arteries with developmental maturation and long-term hypoxia. PLoS ONE. 10(6):33-66. ISSN: 1932-6203. https://dx.doi.org/10.1371%2Fjournal.pone.0130739 [ Links ]
Harmon DL, Swanson CK. 2020. Review: Nutritional regulation of intestinal starch and protein assimilation in ruminants. Animal. 14(1):17-28. ISSN: 2076-2615. https://doi.org/10.1017/S1751731119003136 [ Links ]
Harmon DL. 2009. Understanding starch utilization in the small intestine of cattle. Asian-Australasian Journal of Animal Sciences. 22(7):915-922. ISSN: 1011-2367. https://doi.org/10.5713/ajas.2009.r.08 [ Links ]
Hooijberg EH, Steenkamp G, Buss P, Goddard A. 2017. Method comparison and generation of plasma biochemistry RIs for the White rhinoceros on a point-of-care and wet chemistry analyzer. Veterinary Clinical Pathology. 46(2):287-298. ISSN: 0275-6382. https://doi.org/10.1111/vcp.12490 [ Links ]
Houten SM, Wanders RJA. 2010. A general introduction to the biochemistry of mitocondrial fatty acid β-oxidation. Journal of Inherited Metabolic Disease. 33(1):469-477. ISSN: 0141-8955. http://dx.doi.org/10.1007/s10545-010-9061-2 [ Links ]
Hristov AN, Bannink A, Crompton LA, Huhtanen P, Kreuzer M, McGee M, Nozière P, Reynolds CK, Bayat AR, Yáñez-Ruiz DR, Dijkstra J, Kebreab E, Schwarm A, Shingfield KJ, Yu Z. 2019. Invited review: Nitrogen in ruminant nutrition: A review of measurement techniques. Journal of Dairy Science. 102(1):5811-5852. ISSN: 0022-0302. https://doi.org/10.3168/jds.2018-15829 [ Links ]
Hussain SA, Uppal SK, Randhawa C, Sood NK, Mahajan SK. 2013. Clinical characteristics, hematology, and biochemical analytes of primary omasa impaction in bovines. Turkish Journal of Veterinary and Animal Sciences. 37(1):329-336. ISSN: 1300- 0128. https://doi.org/10.3906/vet-1205-31 [ Links ]
Jiang FG, Lin XY, Yan ZG, Hu ZY, Liu GM, Sun YD, Liu XW, Wang ZH. 2017. Effect of dietary roughage level on chewing activity, ruminal pH, and saliva secretion in lactating Holstein cows. Journal of Dairy Science. 100(4):1-12. ISSN: 0022-0302. https://doi.org/10.3168/jds.2016-11559 [ Links ]
Jin D, Zhao SG, Zheng N, Bu DP, Beckers Y, Wang JQ. 2018. Urea nitrogen induces changes in rumen microbial and host metabolic profiles in dairy cows. Livestock Science. 210(1):104-110. ISSN: 1871-1413. https://doi.org/10.1016/j.livsci.2018.02.011 [ Links ]
Jindal G, Warshel A. 2017. Misunderstanding the preorganization concept can lead to confusions about the origin of enzyme catalysis. Proteins: Structure, Function, and Bioinformatics. 85(12):2-19. ISSN: 1097-0134. https://doi.org/10.1002/prot.25381 [ Links ]
Khezri A, Dayani O, Tahmasbi R. 2016. Effect of increasing levels of wasted date palm on digestion, rumen fermentation and microbial protein synthesis in sheep. Journal of Animal Physiology and Animal Nutrition. 101(1):53-60. ISSN: 0931-2439. https://doi.org/10.1111/jpn.12504 [ Links ]
Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS ONE. 2(1):1112-1126. ISSN: 1932-6203. https://doi.org/10.1371/journal.pone.0047879 [ Links ]
Kohan AB, Wang F, Lo CM, Liu M, Tso P. 2015. ApoA-IV: current and emerging roles in intestinal lipid metabolism, glucose homeostasis, and satiety. Journal of Physiology- Gastrointestinal and Liver Physiolpgy. 308(1):472-481. ISSN: 0193-1857. https://doi.org/10.1152/ajpgi.00098.2014 [ Links ]
Kong F, Liang Y, Légeret B, Beyly-Adriano A, Blangy S, Haslam RP, Napier JA, Beisson F, Peltier G, Li-Beisson Y. 2017. Chlamydomonas carries out fatty acid β-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase. The Plant Journal. 90(1):358-371. ISSN: 0960-7412. http://dx.doi.org/10.1111/tpj.13498 [ Links ]
Kozłowska M, Cieślak A, Jóźwik A, El-Sherbiny M, Stochmal A, Oleszek W, Kowalczyk M, Filipiak F, Szumacher-Strabel M. 2019. The effect of total and individual alfalfa saponins on rumen methane produc. Journal of the Science Food and Agriculture. 100(1):1922-1930. ISSN: 0022-5142. https://doi.org/10.1002/jsfa.10204 [ Links ]
Krause DO, Nagaraja TG, Wright ADG, Callaway TR. 2013. Board-invited review: Rumen microbiology: Leading the way in microbial ecology. Journal Animal Science. 91(1):331-339. ISSN: 1525-3163. https://doi.org/10.2527/jas.2012-5567 [ Links ]
Krehbiel CR. 2014. Invited review: Applied nutrition of ruminants: Fermentation and digestive physiology. The Professional Animal Scientist. 30(1):129-139. ISSN: 1080-7446. https://doi.org/10.15232/S1080-7446(15)30100-5 [ Links ]
Li MM, Sengupta S, Hanigan MD.2019a. Using artificial neural networks to predict pH, ammonia, and volatile fatty acid concentrations in the rumen. Journal of Dairy Science. 102(1):20-32. ISSN: 0022-0302. https://doi.org/10.3168/jds.2018-15964 [ Links ]
Li MMTitgemeyer EC, Hanigan, MD. 2019b. A revised representation of urea and ammonia nitrogen recycling and use in the Molly cow model. Journal of Dairy Science. 102(6):67-88. ISSN: 0022-0302. https://doi.org/10.3168/jds.2018-15947 [ Links ]
Loncke C, Nozière P, Bahloul L, Vernet J, Lapierre H, Sauvant D, Ortigues-Marty I. 2015. Empirical prediction of net splanchnic release of ketogenic nutrients, acetate, butyrate and β-hydroxybutyrate in ruminants: a meta-analysis. Animal. 9(3):449-463. ISSN: 2076-2615. https://doi.org/10.1017/S1751731114002638 [ Links ]
MartineS A-CMF, Van Eunen K, Reijngoud D-J, Bakker BM. 2017. The promiscuous enzyme medium-chain 3-keto-acyl-CoA thiolase triggers a vicious cycle in fatty-acid beta- oxidation. PLoS Computational Biology. 13(4):100-123. ISSN: 1553-734X. https://doi.org/10.1371/journal.pcbi.1005461 [ Links ]
McFadden JW.2020. Review: Lipid biology in the periparturient dairy cow: contemporary perspectives. Animal 14(S1): s165-s175. ISSN: 0968-0004. ISSN: 2076-2615. https://doi.org/10.1017/S1751731119003185 [ Links ]
Menger MF, Nome F. 2019. Interaction vs preorganization in enzyme catalysis. A dispute that calls for resolution. ACS Chemical Biology. 14(1):1386-1392. ISSN: 1554-8929. https://doi.org/10.1021/acschembio.8b01029 [ Links ]
Mikołajczyk K, Pecka-Kiełb E, Zachwieja A. 2019. Impact of the volume and the profile of volatile fatty acids in the rumen fermentation on cow productivity and milk composition. Mljekarstvo. 69(4):222-228. ISSN: 0026-704X. https://doi.org/10.15567/mljekarstvo.2019.0402 [ Links ]
Morita M, Matsumoto S, Okazaki A, Tomita K, Watanabe S, Kawaguchi K, Minato D, Matsuya Y, Shimozawa N, Imanaka T. 2016. A novel method for determining peroxisomal fatty acid β-oxidation. Journal of Inherited Metabolic Disease. 39(1):725-731. ISSN: 0141-8955. http://dx.doi.org/10.1007/s10545-016-9952-y [ Links ]
Moyano JC, López JC, Galván DC, Marini PR, Fischman ML. 2018. Daily variations in protein and energy metabolism during the day in hair sheep in the ecuadorian Amazon Region. Journal of Vetetinaty Science & Technology. 9(2):19-23. ISSN: 2157-7579. https://doi.org/10.4172/2157-7579.1000530 [ Links ]
Norris GH, Jiang C, Ryan J, Porter CM, Blesso CN. 2016. Milk sphingomyelin improves lipid metabolism and alters gut microbiota in high fat diet-fed mice. The Journal of Nutritional Biochemistry. 30(1):93-101. ISSN: 0955-2863.https://doi.org/10.1016/j.jnutbio.2015.12.003 [ Links ]
Nunes-NESI A, Araujo WL, Obata T, Fernie AR. 2013. Regulation of the mitocondrial tricarboxylic acid cycle. Current Opinion in Plant Biology. 16(1):335-343. ISSN: 1369-5266. http://dx.doi.org/10.1016/j.pbi.2013.01.004 [ Links ]
Osorio JH, Barrera LM, Pérez JE. 2015. Comparación del perfil lipídico por sexo y edad en ovinos. Revista de la Facultad de Medicina Veterinaria y de Zootecnia. 62(1):11-19. ISSN: 0120-2952. https://doi.org/10.15446/rfmvz.v62n1.49381 [ Links ]
Panov A, Orynbayeva Z, Vavilin V, Lyakhovich V.2014. Fatty acids in energy metabolism of the central nervous system. BioMed Research International. 20(1):30-42. ISSN: 2414-6133. http://dx.doi.org/10.1155/2014/472459 [ Links ]
Park CJ, Armenia SJ, Shaughnessy MP, Greig CJ, Cowles RA. 2019. Potentiation of serotonin signaling leads to increased carbohydrate and lipid absorption in the murine small intestine. Journal of Pediatric Surgery. 54(1):1245-1249. ISSN: 0022-3468.https://doi.org/10.1016/j.jpedsurg.2019.02.027 [ Links ]
Poher AL, Tschöp MH, Müller TD. 2018. Ghrelin regulation of glucose metabolism. Peptides. 100(1):236-242. ISSN: 0196-9781. https://doi.org/10.1016/j.peptides.2017.12.015 [ Links ]
Pourazad P, Khiaosa-ard R, Qumar M, Wetzels SU, Klevenhusen F, Metzler-Zebeli BU, Zebeli Q. 2016. Transient feeding of a concentrate-rich diet increases the severity of subacute ruminal acidosis in dairy cattle. Journal of Dairy Science. 94(1):726-738. ISSN: 0022-0302. https://doi.org/10.2527/jas.2015-9605 [ Links ]
Prieto ME, Mahecha LL, Angulo AJ, Vargas SJE. 2016. Efecto de la suplementación lipídica sobre ácidos grasos en leche de vaca, énfasis en ácido ruménico. Agronomía Mesoamericana. 27(2):421-437. ISSN: 2215-3608. https://doi.org/10.15517/am.v27i2.22022 [ Links ]
Puppel K, Kuczyńska B. 2016. Metabolic profiles of cow’s blood; a review. Journal of the Science Food and Agriculture. 96(1):4321-4328. ISSN: 0022-5142. https://doi.org/10.1002/jsfa.7779 [ Links ]
Qaid MM, Abdelrahman MM.2016. Role of insulin and other related hormones in energy metabolism-A review. Cogent Food and Agriculture. 2(1):126-142. ISSN: 2331-1932. http://dx.doi.org/10.1080/23311932.2016.1267691 [ Links ]
Qiyu D, Rong Z and Tong F.2019. Review of strategies to promote rumen development in calves. Animals. 9(8):2-15. ISSN: 2076-2615. https://doi.org/10.3390/ani9080490 [ Links ]
Qumar M, Khiaosa-ard R, Pourazad P, Wetzels SU, Klevenhusen F, Kandler W, Aschenbach JR, Zebeli Q.2016. Evidence of in vivo absorption of lactate and modulation of short chain fatty acid absorption from the reticulo-rumen of non lactating cattle fed high concentrate diets. PloS ONE. 11(10):1-15. ISSN: 1932-6203. https://doi.org/10.1371/journal.pone.0164192 [ Links ]
Ramsay JD, Evanoff R, Mealey RH, Simpson EL.2019. The prevalence of elevated γ- glutamyltransferase and sorbitol dehydrogenase activity in racing Thorough breds and their associations with viral infection. Equine Veterinary Journal. 51(1):738-742. ISSN: 0425-1644. https://doi.org/10.1111/evj.13092 [ Links ]
Resende Jr JC, Daniel JLP, Barreto-Vianna ARC, Peixoto JV, Guimarães GC, Costa SF, Lima RF, Meirelles FC. 2019. Determination of volatile fatty acids clearance in intact ruminal digesta. Revista CES Medicina Veterinaria y Zootecnia. 14(1):8-17. ISSN: 1900-9607. http://dx.doi.org/10.21615/cesmvz.14.1.1 [ Links ]
Rostom H, Shine B. 2018. Basic metabolism: proteins. Journal of Basic Science. 30(6):234-240. ISSN: 2448-4997. https://doi.org/10.1016/j.mpsur.2018.01.009 [ Links ]
Rotta PP, Valadares-Filho SC, Detmann E, Costa-Silva LF, Paulino MF, Marcondes MI, Lobo AAG, Villadiego FAC. 2014. Digesta sampling sites and marker methods for estimation of ruminal outflow in bulls fed different proportions of corn silage or sugarcane. Journal of Dairy Science. 92(1):2996-3006. ISSN: 0022-0302. https://doi.org/10.2527/jas.2013-7364 [ Links ]
Schuba J, Südekum KH, Pfeffer E, Jayanegara A. 2017. Excretion of faecal, urinary urea and urinary non-urea nitrogen by four ruminant species as influenced by dietary nitrogen intake: A meta-analysis. Livestock Science. 198(1):82-88. ISSN: 1871-1413. http://dx.doi.org/10.1016/j.livsci.2017.01.017 [ Links ]
Shi F, Wang H, Degen AA, Zhou J, Guo N, Mudassar S, Long R. 2019. Rumen parameters of yaks (Bos grunniens) and indigenous cattle (Bos taurus) grazing on the Qinghai-Tibetan Plateau. Journal of Animal Physiology Animal Nutrition. 103(1):969-976. ISSN: 1439-0396. https://doi.org/10.1111/jpn.13095 [ Links ]
Shi HB, Du Y, Zhang CH, Sun C, He YL, Wu YH, Liu JX, Luo J, Loor JJ. 2018. Fatty acid elongase 5 (ELOVL5) alters the synthesis of long-chain unsaturated fatty acids in goat mammary epithelial cells. Journal Dairy Science. 101(5):4586-4594. ISSN: 0022-0302. https://doi.org/10.3168/jds.2017-14061 [ Links ]
Silva M, Rosani VM, Pinto de Carvalho GG, Vieira PAJ, Alburquerque PML, Pereira L. Campos SF, Fernandes PA, Santana BL, Jeruzia VM, Almeida RLM. 2016. Nitrogen balance, microbial protein synthesis and ingestive behavior of lambs fed diets containing cottonseed cake in substitution of soybean meal semina. Ciências Agrárias. 37(4):2155-2166. ISSN: 2183-041X http://dx.doi.org/10.5433/1679-0359.2016v37n4p2155 [ Links ]
Silva VO, Lopes E, Andrade EF, Sousa RV, Zangeronimo MG, Pereira LJ. 2014. Use of biodiesel co-products (Glycerol) as alternative sources o energy in animal nutrition: a systematic review. Archivos de Medicina Veterinaria. 46(1):111-120. ISSN: 0301-732X. http://dx.doi.org/10.4067/S0301-732X2014000100015 [ Links ]
Song S, Wu J, Zhao S, Casper DP, Zhang L, He B, Lang X, Wang C, Gong X, Wang F, Liu L. 2018. The effect of periodic energy restriction on growth performance, serum biochemical indices, and meat quality in sheep. Journal Animal Science. 96(1):4251-4263. ISSN: 1525-3163. http://dx.doi.org/10.1093/jas/sky299 [ Links ]
Teklebrhan T, Wang R, Wang M, Wen MW, Tan LW, Zhang XM, Ma ZY, Tan ZL. 2020. Effect of dietary corn gluten inclusion on rumen fermentation, microbiota and methane emissions in goats. Animal Feed Science and Technology. 259(1):114-122. ISSN: 0377-8401. https://doi.org/10.1016/j.anifeedsci.2019.114314 [ Links ]
Toral PG, Hervás G, Carreño D, Leskinen H, Belenguer A, Shingfield JK, Frutos F. 2017. In vitro response to EPA, DPA, and DHA: Comparison of effects on ruminal fermentation and biohydrogenation of 18-carbon fatty acids in cows and ewes. Journal of Dairy Science. 100(8):6187-6198. ISSN: 0022-0302. https://doi.org/10.3168/jds.2017-12638 [ Links ]
Toral PG, Monahan FJ, Hervá G, Frutos P, Moloney AP. 2018. Review: Modulating ruminal lipid metabolism to improve the fatty acid composition of meat and milk. Challenges and opportunities. Animal. 12(3):449-463. ISSN: 2076-2615. https://doi.org/10.1017/S1751731118001994 [ Links ]
Tran LV, Malla AM, Kumar S, Tyagi TKA. 2017. Polyunsaturated fatty acids in male ruminant reproduction-A Review. Asian-Australasian Journal of Animal Sciences. 30(5):622-637. ISSN: 1011-2367. https://doi.org/10.5713/ajas.15.1034 [ Links ]
Valdebenito R, Ruminot I, Garrido-Gerter P, Fernández-Moncada I, Forero-Quintero L, Alegría K, Becker HM, Deitmer JW, Barros LF. 2016. Targeting of astrocytic glucosa metabolism by β-hydroxybutyrate. Journal of Cerebral Blood Flow & Metabolism. 36(10):1813-1822. ISSN: 0271-678X. https://doi.org/10.1177/0271678X15613955 [ Links ]
Valente TNP, Lima ES, dos Santos WBR, Cesário AS, Tavares CJ, Fernandes IL, de Freitas MAM. 2016. Ruminal microorganism consideration and protein used in the metabolism of the ruminants: A review. African Journal of Microbiology Research. 10(14):456-562. ISSN: 1996-0808. https://doi.org/10.5897/AJMR2016.7627 [ Links ]
Van Cleef EHCB, Almeida MT, Leal PH, Paschoaloto JR, Filho ESC, Ezequiel JMB. 2018. Effects of partial or total replacement of corn cracked grain with high concentrations of crude glycerin on rumen metabolism of crossbred sheep. Small Ruminant Research. 159(1):45-51. ISSN: 0921-4488. https://doi.org/10.1016/j.smallrumres.2017.12.011 [ Links ]
Vargas JAC. 2019. Función y metabolismo de ácidos grasos en el tejido adiposo y hepático de rumiantes en producción: una revisión. Revista CES Medicina Veterinaria y Zootecnia. 14(2):30-44. ISSN: 1900-9607. http://dx.doi.org/10.21615/cesmvz.14.2.3 [ Links ]
Walther TC, Farese Jr RV. 2012. Lipid droplets and cellular lipid metabolism. Annual Review of Biochemistry. 81(1):687-714. ISSN: 0066-4154. https://doi.org/10.1146/annurev-biochem-061009-102430 [ Links ]
Wallace RJ, Snelling TJ, McCartney CA, Tapio I, Strozzi F.2017. Application of meta‑omics techniques to understand greenhouse gas emissions originating from ruminal metabolism. Genetics Selection Evolution. 49(9):3-14. ISSN: 0999-193X. https://doi.org/10.1186/s12711-017-0285-6 [ Links ]
Wang M, Wang R, Janssen PH, Zhang XM, Sun XZ, Pacheco D, Tan ZL. 2016. Sampling procedure for the measurement of disolved hydrogen and volatile fatty acids in the rumen of dairy cows. Journal Animal Science. 94(1):1159-1169. ISSN: 1525-3163. https://doi.org/10.2527/jas.2015-9658 [ Links ]
Watts JL, Ristow M. 2017. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics. 207(1):413-446. ISSN: 1943-2631. https://doi.org/10.1534/genetics.117.300106 [ Links ]
Witus LS, Netirojjanakul C, Palla KS, Muehl EM, Weng CH, Iavarone AT, Francis MB. 2013. Site-Specific protein transamination using N-Methylpyridinium-4-carboxaldehyde. Journal of the American Chemical Society. 135(1):17223−17229. ISSN: 0002-7863. https://doi.org/10.1021/ja408868a [ Links ]
Yazdi MH, Mirzaei-Alamouti HR, Amanlou H, Mahjoubi E, Nabipour A, Aghaziarati N, Baumgard LH. 2016. Effects of heat stress on metabolism, digestibility, and rumen epithelial characteristics in growing Holstein calves. Journal of Dairy Science. 94(1):77-89. ISSN: 0022-0302. https://doi.org/10.2527/jas.2015-9364 [ Links ]
Yohe TT, Schramm S, White RR, Hanigan MD, Parsons CLM, Tucker HLM, Enger BD, Hardy NR, Daniels KM.2019. Form of calf diet and the rumen. II: Impact on volatile fatty acid absorption. Journal of Dairy Science. 102(9):8502-8512. ISSN: 0022-0302. https://doi.org/10.3168/jds.2019-16450 [ Links ]
Zeng Y, Zeng D, Ni X, Zhu H, Jian P, Zhou Y, Xu S, Lin Y, Li Y, Yin Z, Pan K, Jing B. 2017. Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using illumina MiSeq sequencing revealed high microbial diversity. AMB Express. 7(75):2-10. ISSN: 2191-0855. https://doi.org/10.1186/s13568-017-0378-1 [ Links ]
Zhou H, Meng L, Yin X, Liu Y, Xu G, Wu J, Wu M, Yang L. 2019. Artificial biocatalytic cascade with three enzymes in one pot for asymmetric synthesis of chiral unnatural amino acids. European Journal Organic Chemistry. 38(1):6470-6477. ISSN: 1099-0690. https://doi.org/10.1002/ejoc.201900828 [ Links ]
Received: April 02, 2020; Accepted: July 10, 2020











 texto en
texto en