Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Abanico veterinario
On-line version ISSN 2448-6132Print version ISSN 2007-428X
Abanico vet vol.10 Tepic Jan./Dec. 2020 Epub Mar 02, 2021
https://doi.org/10.21929/abavet2020.10
Article original
Genetic diversity and population structure of Yucatan black hairless pig using SNP50K chip
1Posgrado en Ciencias Biológico Agropecuarias, Universidad Autónoma de Nayarit. México.
2Departamento de Genética y Bioestadística, Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México. México.
3Asociación Mexicana de Criadores de Cerdos de Origen Ibérico Yucatán, A. C. México.
4Corporación Colombiana de Investigación Agropecuaria-AGROSAVIA, Centro de Investigación Tibaittá, Colombia.
5Instituto Tecnológico de Conkal, Yucatán. México.
In the present study, the Population structure and genetic diversity of 104 Yucatan black hairless pigs (YBH) and 8 Duroc breeds were by using an SNP50K chip characterized. The population structure was obtained, as well as the calculation of Principal Component Analysis (PCA), Minor Allele Frequency (MAF), observed heterozygosity (oH), consanguinity (F), Fixation index of individuals within subpopulations (Fis), the t (outcrossing rate or alogamia index) was made, also the association analysis to identify SNP with population differences. The genetic component of Duroc in YBH subpopulations is low, from 0.00363 to 0.03532, THUS, IT WAS OBSERVED (appreciating) a subpopulation with greater genetic diversity and lower values of F and Fis, as well as higher oH and t. SNPs were identified (p<1.213E-50 to p< 6.4E-20), associated with genes and biological processes. Genes EHF, DST, PDE8A, FOXA1 and VCL are related to epithelial cell differentiation, morphogenesis, and development of epithelium. Other 30 SNPs are to nutrient metabolism related, 23 SNPs to nutrient transport, 11 SNPs to immunity, 10 SNPs to muscle, skeletal and embryonic, and 7 SNPs to synapses and receptors. YBH is distant from Duroc with different population structure and genetic diversity with different genes that involve important biological processes.
Keywords: genetic resources; SNP; genetic diversity; population structure; creole pig
La estructura poblacional y diversidad genética de 104 cerdos negros lampiños de Yucatán (NLY) y ocho de raza Duroc fueron caracterizados usando un chip SNP50K. Se obtuvo la estructura poblacional, se calculó un análisis de componentes principales (ACP), menor alelo frecuencia (MAF), Heterocigosidad observada (Ho), Consanguinidad (F), Índice de Fijación de individuos en subpoblaciones (Fis), índice de alogamia (t) y análisis de asociación para identificar SNP diferentes entre poblaciones. Según el análisis Admixture la población NLY se estructura en tres subpoblaciones. El componente genético de Duroc en subpoblaciones NLY es bajo de 0.0036 a 0.0353, apreciándose una subpoblación con mayor diversidad genética, con valores más bajos de F, Fis y mayor Ho y t. Se identificaron SNP (p<1.21E-50 a p< 6.4E-20), asociados con genes y procesos biológicos. Genes EHF, DST, PDE8A, FOXA1 y VCL relacionados con la diferenciación de células epiteliales, la morfogénesis y desarrollo del epitelio. Otros 30 SNPs relacionados con el metabolismo de nutrientes, 23 SNPs en transporte de nutrientes, 11 SNPs a inmunidad, 10 SNPs a músculo, esqueleto y embrionario, y siete SNPs a sinapsis y receptores. NLY está distante de Duroc con diferente estructura poblacional y diversidad genética, con diferentes genes que implican procesos biológicos importantes.
Palabras claves: recursos genéticos; SNP; diversidad genética; estructura poblacional y cerdo criollo
INTRODUCTION
Since the arrival of pigs in America in the 16th century and their distribution throughout the new world, whether in natural or artificial selection, it has shaped the diversity of today’s populations. Burgos-Paz et al. (2013) described how the environment influenced the phenotypic differences between pigs in the highlands of Peru, in relation to those that inhabit lowland or tropical. Taking into account the wide range of climates in America, and particularly in the coasts, some pig populations are growing in size and relevance to human communities. This is the case of Yucatan hairless black pigs (YBH); this breed of pig has phenotypic peculiarities: black, hairless, flawless skin, black hoof and straight snout with an important participation in the food security of rural human populations (Lemus y Alonso, 2005).
The FAO Animal Diversity Information System (DAD-IS, 2020), considered that this breed was in danger of extinction, without a conservation program, and it is a valuable animal genetic resource that can contribute to food security in rural communities; as well as a reservoir of genetic diversity (Lemus-Flores et al., 2001; Scarpa et al., 2003). In addition, the status of 38% of pig breeds worldwide is unknown (FAO, 2019).
This hairless biotype comes from Iberian pigs (Sus mediterraneus), of African origin, introduced in all southern European regions (Benítez y Sánchez, 2001). They were later introduced to America on Christopher Columbus’ second voyage in 1493 (Ogata, 2019). YBH pigs are a genetic variation of Creole pigs (Su et al., 2014) and are divided into two genetic lines; one located in populations in the Gulf of Mexico and the other population is present on the Mexican Pacific coast (Lemus-Flores et al., 2001); it is called hairless Mexican, hairless or tropical hairless. They were at risk of extinction due to unplanned crossings with commercial lines of lean genotypes. Despite the fact that YBH is widely distributed on the Yucatan peninsula in Mexico, with low to medium-sized technical management. Currently, some pig farmers have found a valuable way to raise it, but the available information is limited in the population structure or lineage.
To obtain a breeding program in order, a large population of YBH was using a commercial SNP50K chip evaluated, to estimate the diversity and the actual population structure, as well as their genetic relationships for the future selection of feet of distant commercial breeds.
MATERIAL AND METHODS
Animals and genotype análisis
From a total population of 560 hairless black pigs from Yucatan (YBH) from 49 farms, located from Mérida to Tizimín in Yucatan state, Mexico; 104 breeding adults from 2 to 3 years old (17 boars and 87 sows) were selected, considering phenotypically characteristics of hairlessness (hairless), black skin, without spots, black hoof and straight snout. In addition, information about the origin of the sows or boars was used to reduce any relationship between the samples. In addition, eight Duroc sows were sampled as a reference population and they were used to assess introgression in Yucatan pigs.
This study has registration SIP18-076 from the Autonomous University of Nayarit and an agreement with the Yucatan Science and Technology Park. For the collection of blood samples, the recommendations of Official Standards NOM-051-ZOO-1995, on human treatment of animals, and NOM-062-ZOO-1999, of the technical specifications for production, care and testing were followed use of laboratory animals. The extraction and genotyping of genomic DNA from blood samples was carried out at NEOGEN (www.neogen.com). For SNP genotyping, porcine GGP-50K was used, which identifies 50,967 SNPs (GeneSeek Genomic Profiler Porcine).
Quality control of SNP genotypes (Simple nucleotide polymorphism)
SNP data quality control was performed using PLINK v1.9 (Purcell et al., 2007). The SNP with polymorphisms <0.10 and MAF (Frequency of minor allele) <0.01, if excluded. The 42840 SNP were retained for further analysis of the structure of the population and genetic diversity.
Statistical análisis
Analysis of the population structure
First, an analysis of the population structure was to identify the subpopulations in YBH pigs performed with the Admixture 1.3 software (Alexander et al., 2009). Principal Component Analysis (PCA) was obtained with PLINK v2.1 (Chang et al., 2015) and a graph was constructed using Minitab v15 software to visualize the genetic distances between the YBH subpopulations and the Duroc breed.
Genetical diversity
For each subpopulation of pig YBH and Duroc, the minor allelic frequency (MAF), the observed heterozygosity (oH), consanguinity (F) and the index of fixation of individuals in the subpopulations (Fis) were calculated using the PLINK v1.9 program (Pursell et al., 2007). The t (crossing rate or allogamy index) was according to Weir (1990)calculated. To compare the YBH and Duroc pig sub-populations, the analysis of variance of a single classification criterion was used (SPSS v20, 2011).
Candidate SNP regions
With PLINK v1.9 (Pursell et al., 2007) for the entire population of pig YBH vs Duroc, the association was calculated to identify SNPs with differences. The genetic annotations in the candidate regions were using the preliminary annotation of assembly 10.2 obtained, provided by e-ensembl (Groenen et al., 2012). The over representation of Gene Ontology (GO) categories was determined using the Gene Ontology database (Ashburner et al., 2000).
RESULTS
Population structure
The first population structure was performed with a 10-fold cross-validation, to choose the best K value; the K=4 value presented the smallest cross-validation error (0.584). According to the mixture analysis, the YBH population is in three subpopulations structured (Table 1).
Table 1 Composition of the cluster predicted by ADMIXTURE (K4)
| DUR | YUC1 | YUC2 | YUC3 | |
|---|---|---|---|---|
| DUR | 0.99997 | 0.03532 | 0.00363 | 0.02726 |
| YUC1 | 0.00001 | 0.69764 | 0.18677 | 0.18429 |
| YUC2 | 0.00001 | 0.08993 | 0.64234 | 0.07703 |
| YUC3 | 0.00001 | 0.17711 | 0.16726 | 0.71142 |
YUC1, YUC2 and YUC3 sub-populations of the hairless black pig of Yucatan. DUR Duroc breed population. The values per column of each breed indicate the proportions of other breeds.
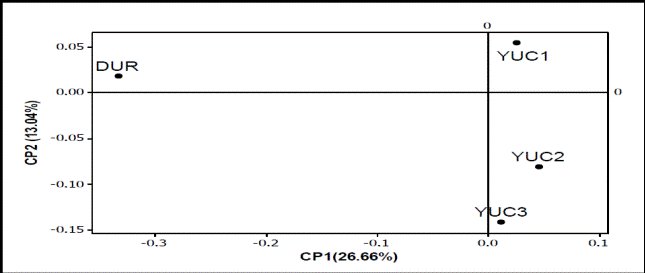
With the ACP, YUC2 subpopulation is the most distant from the Duroc breed, with YUC1 and YUC3 closer because they share more Duroc genetic component (graph 1).
Genetical diversity
Positive values of Fis indicate consanguinity; is greater if it approaches 1; corresponds to the general reduction in heterozygosity observed in relation to that expected in its population; in the YUC3 subpopulation, the Fis value is lower, F and oH are higher, indicating greater genetic diversity (Table 2).
Table 2 Genetic diversity of black hairless pigs of Yucatan and Duroc breed
| YUC1 | YUC2 | YUC3 | Duroc | eem | |
|---|---|---|---|---|---|
| Samples | 70 | 14 | 20 | 8 | |
| MAF Mean | 0.260ª | 0.216c | 0.247b | 0.202d | 0.025 |
| Consanguinity (F) | 0.039ª | 0.067a | -0.006 b | 0.079a | 0.011 |
| oH | 0.328b | 0.305b | 0.359 a | 0.301b | 0.011 |
| Fis | 0.079ª | 0.142 a | -0.007 b | 0.158a | 0.021 |
| Allogamy index (t) | 0.870b | 0.782 b | 1.0278 a | 0.729b | 0.027 |
MAF, minor allele’s frequency. oH, observed heterozygosity. Fis, sub-population fixation index. eem, mean standard error. Different letters in the rows indicate statistical different between populations (ANOVA, p<0.05).
A population approaches random mating if the value of t approaches 1, when it is greater than 1, there is an excess of heterozygotes and when the value is zero, all individuals are homozygous; in the YUC3 subpopulation, it approaches 1, being lower in the other YUC1, YUC2 and Duroc subpopulations.
Candidate SNP regions
In the analysis of association between the entire population of YBN vs Duroc to identify differences, 226 SNPs were identified with values of p <1.21E-50 ap <6.4E-20, of which only 93 SNPs were associated with genes and biological processes (Table 3).
Table 3 Biological processes, genes and information from SNPs that showed the greatest differentiation between YBH vs Duroc
| Biological process | Chromosome | Variants | Identified genes |
|---|---|---|---|
| Epithelial cell differentiation, morphogenesis and epithelial development | 2 | rs81223208 | EHF |
| 7 | rs80830437, rs331746636, rs81222725, rs81398056, rs325625775 | DST, PDEA8A, FOXA1 | |
| 14 | rs80785304, rs345768654 | VCL | |
| Nutrient metabolism | 2 | rs713429023 | PDHX |
| 3 | rs81317284 | ST6GAL2 | |
| 6 | rs81390019, rs81390069, rs81390070, rs81390137, rs81285728, rs81317489, rs81226716, rs81318326, rs81475823 | PABPC4, HPCAL4, MFSD2A, MC2R, MPPE1, IMPA2, PTPN2 | |
| 7 | rs80793059, rs342597254, rs80868794, rs80837023, rs80951652, rs80986501, rs80845345, rs80850402 | GCLC, ADAMTSL3, HOMER2, UNC45A | |
| 13 | rs81448371 | PDIA5 | |
| 14 | rs339061874, rs328957349, rs345309524, rs80889570, rs80895748, rs80897302 | CFAP70, CHCHD1, ADK, DUSP13 | |
| 15 | rs81241812 | ACSL1 | |
| X | rs327444342, rs322147119, rs81474003 | FAM58A, BRCC3 | |
| Nutrient transport | 1 | rs328115005 | VPS39 |
| 6 | rs81389915, rs329679425, rs81389921, rs81251860, rs81389936, rs81389948, rs81389955 , rs81389959, rs81262099, rs81211910, rs81390112 | MACF1,TRIT1 | |
| 12 | rs81261131 | PITPNC1 | |
| 14 | rs81451083, rs81451108 | MICU1, CAMK2G | |
| 15 | rs343808632 | TRAK2 | |
| 18 | rs81471732 | SLC13A4 | |
| x | rs326399484, rs337683495, rs81473903, | VMA21, PASD1, ZNF185, rs81473906, rs80784223, rs80910586 | |
| Immunity | 6 | rs341367004, rs81306790 | RNMT |
| 7 | rs81398013, rs80837723, rs80805016, rs80976160, s80849899 | IL6 | |
| 15 | CTLA4 | ||
| X | rs328334089, rs327024720, rs336767148 | IL1RAPL2 | |
| Muscle, skeletal and embryonic development. | 4 | rs326729657 | ARNT |
| 6 | rs81389986, rs339432830 | BMP8B, MYOM1 | |
| 7 | rs80816179, rs81398046 | TM6SF1, CPEB1 | |
| 8 | rs81476832 | TSPAN5 | |
| 9 | rs81305287 | PRRC2C | |
| 14 | rs327184000 | P4HA1 | |
| 15 | rs80949190 | SATB2 | |
| x | rs81474001 | VBP1 | |
| Synapses and receptors | 3 | rs81370102 | SLC5A7 |
| 6 | rs81337627 | GRIK3 | |
| 18 | rs322407819 | GRM8 | |
| x | rs330548482, rs322056532, rs325753884, rs80918182 | GABRA3, GABRQ |
Considering the number of different SNPs between YBH and Duroc, on chromosomes 6, 7, X and 14, this is where the highest number was identified (Table 4).
DISCUSSION
The 104 YBH samples chosen for the study were hairless muzzles, flawless black skin, black hull and straight foci to avoid phenotypic variations. The ADMIXTURE analysis subdivides them into three subpopulations. The introgression of the Duroc breed is very low in the three identified YBH subpopulations (YUC1, YUC2 and YUC3) from 0.00363 to 0.03532. It is similar to that of the Pampa Rocha pig from Uruguay (Montenegro et al., 2015), inferior to the others Creole pigs in America, in which the Duroc component has an average of 0.15 and ranges from 0.00 (US Yucatan) to 0.45 (Moura Brasil) (Burgos-Paz et al., 2013). The hairless pig in the Mexican Pacific has 0.20 of the Duroc component (Lemus et al., 2001).
The separation of three subpopulations in the YBH and the low introgression of Duroc coincide with that observed by Burgos-Paz et al. (2013). Modern pigs in the Americas are the result of many independent colonization events, but also to environmental challenges; as these YBH pigs are geographically isolated, far from commercial and hairless pigs, located on the Pacific and Gulf coasts. There is no evidence of an artificial breeding or breeding program with commercial breeds on the Yucatan peninsula.
There is a difference in genetic diversity between YBH subpopulations; in YUC3 it is taller, approaching random mating. It is likely that individuals from YUC1, YUC2 and Duroc show matings from individuals with greater genetic proximity in each subpopulation, which caused an increase in F and Fis. With the information generated, it is possible to program the mating in the subpopulations, generate more genetic diversity and avoid the request for variability, as occurs in Iberian populations (Esteve et al., 2013). It is important to consider the statement by Yang et al. (2017)), indicating that populations with high Fis, normally present low diversity of haplotypes.
By identifying 93 SNPs that are different between populations of YBH vs Duroc, according to their extreme allele frequencies, higher frequency in Duroc and lower in YBN, we can use them as markers. According to Yan et al. (2017), domestication and artificial selection, gave rise to a wide range of phenotypes among domestic pig breeds, which differ from their wild relatives; and that these are to behavior, body size, fertility, ability to move and adapt to food provided by humans related. Therefore, it is important to detect genetic loci, which may be involved in the transition from wild to domestic.
Considering that the American pigs are part of Iberian origin (Burgos-Paz et al., 2013), YBH is phenotypically similar to the Iberian hairless, so that their coat is predominantly black and there are no white pigs. However, as Ramírez et al. (2015), there were no SNPs related to the MC1R gene, which would allow considering the red or black color of the layer. Eight SNPs associated with epithelial cell differentiation, morphogenesis and epithelial development were identified; with five genes that could be studied as candidates for the hairless phenotype.
This study does not coincide with Su et al. (2014), which proposes the BAMB1 gene as a strong hairless candidate; this gene was not different between the YBH and Duroc populations (p <0.35), with very similar allele frequencies. With Burgos-Paz et al. (2013) coincides with the PDE family genes, they report that in American Creole pigs the PDE10A and PDE11A genes are identified; the PDE8A gene is reported here. Esteve et al. (2013) in hairless Iberian pigs reports the FOXP1 gene as a candidate gene involved in the differentiation of epithelial cells, keratinization and the formation of hair follicles. In this study, the FOXA1 gene was identified, which is from the same family, also identified by Yang et al. (2017), which associates it with European pigs and relates it to the metabolism of proteins, glucose or fatty acids. For lipid metabolism Esteve et al. (2013)report eight candidate genes, not found in the PDHX, MFSD2A and ACSL1 report that were in this study identified; but for an immune response, these researchers coincide on the IL1RAPL2 gene, located on the X chromosome and IL6 on chromosome 7. On chromosome 6, the largest number of SNPs (27), involved in most biological functions, was identified. In this chromosome, for the transport of nutrients, ten SNPs were identified, for the MACF1 gene (cross-linking factor 1 of the microtubule-actin), identified in humans and mice as essential for embryonic development, to maintain the neuronal system and the integrity of the skin (Kang, et al., 2020).
For muscle and skeletal development, the SNP BMP8B has been reported as an important gene for embryonic growth and development characteristics (Xiu-Kai et al., 2013; Ying et al., 2000). For synapses and receptors, four genes related to brain and nervous disorders in humans have been identified what an opportunity for this pig to use it as a biomedical model. The GRM8 gene has also been reported by Kwonm et al. (2019) in Yucatan miniature pig, associated with nervous diseases in humans; similarly, the GABRA3 gene is associated with epileptic diseases (Niturad et al., 2017), the SCL5A7 gene is associated with myasthenic congenital syndrome type 20 (Pardal-Fernández et al., 2018) and GRIK3 in neurophysiological processes.
The high number of SNPs identified as different between YBH and Duroc can be attributed to the lack of selection and crossing with the Duroc breed. Although further studies are needed to validate the role of the identified genes, these findings confirm that domestication and evolution are different between Duroc and YBH pigs.
CONCLUSION
In this first study with SNP50K in YBH pigs, the genetic difference of this pig is in comparison to the Duroc pig identified, providing useful genetic information for the conservation of this local genetic resource. YBH is far from Duroc, with a different population structure, genetic diversity and different genes involving important biological processes, which can be useful in the racial selection and differentiation program.
Suplementary information
Tabla A. List of 93 SNPs and genes associated with biological processes.
ACKNOWLEDGMENTS
This research was funded by the Secretariat for Research, Innovation and Higher Education, Mérida, Yucatán, Mexico.
REFERENCES
Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 19:1655-1664. https://doi.org/10.1101/gr.094052.109 [ Links ]
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 25(1):25-29. https://doi.org/10.1038/75556 [ Links ]
Benítez OW, Sánchez DM. 2001. Los cerdos criollos en América Latina. En: FAO (ed.) Los cerdos locales en los sistemas tradicionales de producción. Estudio FAO Producción y Sanidad Animal. Pp.13-35. http://www.fao.org/3/a-y2292s.pdf [ Links ]
Burgos-PAZ W, Souza CA, Megens HJ, Ramayo-Caldas Y, Melo M, Lemus-Flores C, Caal E, Soto HW, Martínez R, Álvarez LA. 2013. Porcine colonization of the Americas: a 60k SNP story. Heredity. 110:321-330. https://www.nature.com/articles/hdy2012109 [ Links ]
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 4:7. https://academic.oup.com/gigascience/article/4/1/s13742-015-0047-8/2707533 [ Links ]
DAD-IS. 2020. Sistema de Información sobre la Diversidad de los Animales Domésticos (DAD-IS@fao.org). FAOIn: http://www.fao.org/dad-is/browse-by-country-and-species/es/ [ Links ]
Esteve Codina A, Paudel Y, Ferretti L, Raineiri E. 2013. Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics. 14:148- https://doi.org/10.1186/1471-2164-14-148 [ Links ]
FAO. Food and Agriculture Organization. 2019. Domestic Animal Diversity Information System (DADIS). Retrieved March 31, 2020, from Retrieved March 31, 2020, from http://www.fao.org/dad-is/regional-national-nodes/efabis/en/ [ Links ]
Groenen M, Archibald A, Uenishi H, Tuggle C, Takeuchi Y, Rothschild M. 2012. Pig genomes provide insight into porcine demography and evolution. Nature. 491: 393-398. https://www.ensembl.org/Sus_scrofa/Info/Index [ Links ]
Kang L, Liu Y, Jin Y, Li M, Song J, Zhang Y, Zhang Y and Yang Y. 2020 Mutations of MACF1, Encoding Microtubule-Actin Crosslinking-Factor 1, Cause Spectraplakinopathy. Frontiers in Neurology. 10:1335. https://doi.org/10.3389/fneur.2019.01335 [ Links ]
Kwon DJ, Lee YS, Shin D, Won KH, Song KD. 2019. Genome analysis of Yucatan miniature pigs to assess their potential as biomedical model animals. Asian-Australas J Anim Sci. 32(2):290‐296. doi:10.5713/ajas.18.0170 [ Links ]
Lemus C, Alonso ML. 2005. El cerdo Pelón Mexicano y otros cerdos criollos. 1ª Edición. Editorial Universitaria. Universidad Autónoma de Nayarit. México. Pp. 251. https://www.amazon.com.mx/cerdo-pel%C3%B3n-mexicano-cerdos-criollos-ebook/dp/B0153G544M [ Links ]
Lemus-Flores C, Ulloa-Arvizu R, Ramos-Kuri M, Estrada FJ, Alonso RA. 2001. Genetic analysis of Mexican hairless pig populations. Journal Animal Science. 79:3021-3026. https://doi.org/10.2527/2001.79123021x [ Links ]
Ogata N. 2019. 1519, Hernán Cortés y el Cerdo en México. Diversidad Biológica y Cultural Trópico Americano. Centro de Investigaciones Tropicales (CITRO), Universidad Veracruzana. Disponible: http://etnoecologia.uv.mx/diversidad_biocultural/cerdo-pelon- mexicano/ [ Links ]
Montenegro M, Llambí S, Castro G, Barlocco N, Vadell A, Landi V, Delgado JV, Martínez A. 2015. Genetic characterization of Uruguayan Pampa Rocha pigs with microsatellite markers. Genetics and Molecular Biology. 38(1):48-54. https://doi.org/10.1590/S1415-475738120140146 [ Links ]
Niturad C E, Lev D, Vera M, Crzewska A, Schubert J, Lerman-Sagie T, Kroes H, Oegema R, Traverso M, Specchio N, Lassota M, Chelly J, Bennett-Back O, Carmi N, Koffler-Brill T, Lacomino M, Trivisano M, Capovilla G, Striano P, Nawara M, Rzonca S, Fischer U, Bienek M, Jensen C, Hu H, Thiele H, Altmüller J, Krause R, May P, Becker F, Balling R, Biskup S, Haas SA, Nürnberg P, Van Gassen KLI, Lerche H, Zara F, Maljevic S, Leshinsky-Silver E. 2017. GABRA3 variants are associated with epileptic seizures, encephalopathy and dysmorphic features. Brain. 140(11):2879-2894. https://doi.org/10.1093/brain/awx236https://academic.oup.com/brain/article/140/11/2879/4372140 [ Links ]
NOM-051-ZOO-1995. Norma Oficial Mexicana NOM-051-ZOO-1995. Trato humanitario en la movilización de animales. Pp 23. http://publico.senasica.gob.mx/?doc=531 [ Links ]
NOM-062-ZOO-1999. NORMA Oficial Mexicana NOM-062-ZOO-1999. Especificaciones técnicas para la producción, cuidado y uso de los animales de laboratorio. Pp 58. http://publico.senasica.gob.mx/?doc=743 [ Links ]
Pardal-Fernández J M, Carrascosa-Romero M C, Álvarez S, Medina-Monzón M C, Caamaño M B, de Cabo C. 2018. A new severe mutation in the SLC5A7 gene related to congenital myasthenic syndrome type 20. Neuromuscular Disorders. 28(10):881‐884. https://doi.org/10.1016/j.nmd.2018.06.020 [ Links ]
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal Human Genetics. 81(3):559-575. https://doi.org/10.1086/519795. https://www.cell.com/ajhg/fulltext/S0002-9297(07)61352-4 [ Links ]
Ramírez O, Burgos-Paz W, Casas E, Ballester M, Bianco E, Olalde I, Santpere G, Novella V, Gut M, Lalueza C, Saña M, Pérez-Enciso M. 2015. Genome data from a sixteenth century pig illuminate modern breed relationships. Heredity. 114:175-184. https://www.nature.com/articles/hdy201481 [ Links ]
Scarpa R, Drucker AG, Anderson S, Ferraes-Ehuan N, Gómez V, Risopatrón CR, Rubio-Leonel O. 2003. Valuing genetic resources in peasant economies: the case of ‘hairless’ creole pigs in Yucatan. Ecological Economics. 45:427-443. https://doi.org/10.1016/S0921-8009(03)00095-8 [ Links ]
SPSS. Statistical Package for the Social Sciences. 2011. IBM® SPSS® Statistics v20 for Windows: advanced statistic release. SPSS, Chicago. USA. http://public.dhe.ibm.com/software/analytics/spss/documentation/statistics/20.0/es/client/Manuals/IBM_SPSS_Statistics_Base.pdf [ Links ]
Su Y, Long Y, Liao X, Ai H, Zhang Z, Yang B, Xiao S, Tang J, Xin W, Huang L, Ren J, Ding N. 2014. Detection of genomic signatures for pig hairlessness using high-density SNP data. Frontiers of Agricultural Science and Engineering. 1(4):307-313. https://doi.org/10.15302/J-FASE-2014039 [ Links ]
Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38:1358-1370. https://doi.org/10.2307/2408641 [ Links ]
Xiu-Kai C, Jing W, Xian-Yong L, Chu-Zhao L, Chun-Lei Z, Xing-Lei Q, Hong C. 2013. Genetic variants in BMP8B gene are associated with growth traits in Chinese native cattle. Gene. 532(1):115-120. https://doi.org/10.1016/j.gene.2013.09.059 [ Links ]
Yang B, Cui L, Perez-Enciso M, Traspov A, Crooijmans R, Zinovieva N, Schook L, Archibald A. 2017. Genome-wide SNP data unveils the globalization of domesticated pigs. Genetics Selection Evolution. 49:71-85. https://doi.org/10.1186/s12711-017-0345-y [ Links ]
Ying Y, Liu X M, Marble A, Lawson K A, Zhao G Q. 2000. Requirement of Bmp8 for the Generation of Primordial Germ Cells in the Mouse. Molecular Endocrinology. 14(7):1053-1063. https://doi.org/10.1210/mend.14.7.0479 [ Links ]
Received: February 05, 2020; Accepted: June 03, 2020











 text in
text in