Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Abanico veterinario
versión On-line ISSN 2448-6132versión impresa ISSN 2007-428X
Abanico vet vol.9 Tepic 2019 Epub 30-Jul-2021
https://doi.org/10.21929/abavet2019.922
Original articles
Africanization of colonies of Apis mellifera L. (Hymenoptera:Apidae), present in the mitochondrial DNA
1Universidad Autónoma de Tamaulipas, Facultad de Medicina Veterinaria y Zootecnia “Dr. Norberto Treviño Zapata”. Tamaulipas, México. asilvac10b@gmail.com, rigoberto62@hotmail.com
2Universidad Autónoma de Tamaulipas, Facultad de Ingeniería y Ciencias. Centro Universitario Adolfo López Mateos. Tamaulipas, México. jmartinez@docentes.uat.edu.mx
3Universidad de Guadalajara, Centro Universitario del Sur. Jalisco, México. ecienfue@docentes.uat.edu.mx
4Instituto Politécnico Nacional, Centro de Biotecnología Genómica. Reynosa, Tamaulipas, México. joset@cusur.udg.mx gparra@ipn.mx
Mexican beekeeping is an activity associated with producers of low income whose products are intended for export. The present study was conducted to assess the degree of africanization of the commercial apiaries through the presence of African genes in deoxyribonucleic acid (DNA) mitochondrial of honeybees (Apis mellifera L.). We collected bees from commercial apiaries that were kept in alcohol ethanol until the DNA extraction. The Retrieved DNA was frozen at - 20 °C until use. 50 µg that was digested PCR product were used for the PCR-RFLP and incubated for 16 hours and the products of digestion were separated by electrophoresis in an agarose 1% solution TBE 1X gel. The staining of the gels was performed with ethidium bromide. A pattern of two fragments (194 and 291 pb) was visualized corresponding to the mitotype European (E) and a fragment only undigested 485 PB which corresponds to the African mitotype. The population of Hardy-Weinberg are not in balance, there is deficit of heterozygotes.
Keywords: Africanization; PCR; Norther Mexico
La apicultura es una actividad asociada a productores de bajos ingresos cuyos productos están destinados a la exportación. El presente estudio se realizó para evaluar el grado de africanización de los apiarios comerciales a través de la presencia de genes africanos en el ácido desoxirribonucleico (ADN) mitocondrial de abejas melíferas (Apis mellifera L.). Se colectaron abejas de apiarios comerciales, que se mantuvieron en alcohol etanol hasta la extracción del ADN. El ADN obtenido se congeló a -20° C hasta su utilización. Para la PCR-RFLP se utilizaron 50 µg de producto de PCR que fue digerido e incubado por 16 horas y los productos de la digestión fueron separados por electroforesis en un gel de agarosa 1% en solución TBE 1X. La tinción de los geles se realizó con bromuro de etidio. Se visualizó un patrón de dos fragmentos (194 y 291 pb) correspondiente al mitotipo Europeo (E) y un fragmento único sin digerir de 485 pb que corresponde al mitotipo africano. El porcentaje de africanización de las poblaciones fue de 69.9% con base en el análisis polimórfico de ADN mitocondrial, confirmando la dominancia de genes africanizados. Las poblaciones no están en equilibrio de Hardy-Weinberg, porque existe déficit de heterocigotos.
Palabras Clave: Africanización; PCR; Norte de México
INTRODUCTION
The state of Tamaulipas has many multiflora resources that could be used for the development of beekeeping, particularly in the Sierra Madre Oriental (Villegas et al., 2003); in addition, in the center of the state there is an important citrus area (SDR, 2017). However, the africanization of the nuclei has led to the decrease in honey production; also, diseases, parasitosis, lack of training of producers and climatic phenomena have also diminished the productive potential of bees (Guzmán-Novoa et al., 2004; Medina- Flores et al., 2015; Urbina-Romero et al., 2019). Similarly, the introduction of new pests such as the hive beetle (Aethina tumida), recently reported in the country by SAGARPA (2007).
The arrival of Africanized bees in 1987, brought significant changes in the way of beekeeping in the country; these hybrids inherited undesirable characteristics from their African parents (Apis mellifera scutellata), such as defensive behavior, the tendency to swarm and migration (Medina-Flores et al., 2015; Urbina-Romero et al., 2019). The direct consequences of this defensive behavior were the abandonment of the activity of producers, reduction of the number of hives, increase in production costs due to the acquisition of protective equipment and the periodic exchange of fertilized queens (SAGARPA, 2016).
However, Africanized bees have desirable characteristics for apicultural activity, including adaptation to tropical climates that results in rapid population growth and resistance to certain diseases and parasitosis (Vázquez-Castro et al., 2006; Medina -Flores et al., 2015).
On the other hand, the polymerase chain reaction (PCR), which is associated with restriction fragment length polymorphism (RFLP), which are a versatile, fast and low-cost tool to detect single nucleotide polymorphisms (SNPs) of genes associated with production characteristics; as well as to evaluate the genetic variability between and within populations (Sifuentes et al., 2006). It has been proven that polymorphism in nuclear DNA (DNA) and mitochondrial DNA (DNAm) are very useful molecular markers in the study of population genetics (Clarke et al., 2001).
Likewise, the DNA polymorphism was used to discriminate groups of bees belonging to the African and European subspecies (Esquivel-Rojas et al., 2015). On the other hand, Medina-Flores et al., (2015) found in a study conducted in Zacatecas, Mexico, that the differences in the frequencies of African and European morphotypes were significant, both intraregionally and between the three regions.
The present study aimed to characterize Apis mellifera bee populations of commercial apiaries in the state of Tamaulipas, using molecular analysis techniques to identify DNAm variability and the presence of European and African genes.
MATERIAL AND METHODS
103 samples were collected from commercial apiaries in the state of Tamaulipas; producers replace their queens approximately every two years (SAGARPA, 2016), with certified queen bees (not Africanized); Some resort to artificial feeding of bees during the cold and dry seasons, with products such as fructose syrup and soy cake.
The samples (bees) were kept in 95 % absolute ethanol alcohol at room temperature until DNA extraction. The complete DNA extraction was carried out in the Molecular Biology Laboratory of the Faculty of Veterinary Medicine and Zootechnics of the Autonomous University of Tamaulipas, according to the protocol proposed by Crozier and Crozier (1993). For the extraction of DNA, four bees were frozen with liquid nitrogen (N2), macerated in sterile mortar and the samples were digested at 55 °C for 16 hours in 295 µL of Master-Mix solution (Proteinase K, RNase A Solution, EDTA 0.5 M, pH 8.0; Nuclei Lysis Solution). To purify the DNA, the 1.5 mL Eppendorf® tube was added to the sample, 250 µL of Wizard® SV Lysys Buffer, centrifuged at 13000 Xg for one minute; was transferred to the collecting tube with a mini-column with filter, 650 µL of Wizard® SV Wash Solution (four times) was added, centrifuging at 13000 Xg for one minute. Subsequently, the minicolumn tube was discarded and the filter was placed with the DNA in a new 1.5 ml Eppendorf® tube, 250 µL of deionized sterile nuclease-free H2O was added by incubating for two minutes at room temperature. The sample was centrifuged at 13000 Xg for two minutes and the DNA obtained was frozen at -20 °C until use.
The PCR was developed according to the protocol described by Esquivel-Rojas et al. (2015), but with 100 ng of DNA, 50 nm of each initiator and 25 µL of Mastermix®, which includes 1 U of Taq polymerase and 10 µL of sterile nuclease-free H2O.
The 485 bp region of the Cytochrome b gene was amplified using the Cytb-b F initiators: TAT GTA CTA CCA TGA GGA CAA ATA TC and Cytb-R ATT ACA CCT CCT AAT TTA TTA GGA AT. The mixture was denatured at 94 °C for one minute, followed by 30 cycles, 94 °C for one minute, 60 °C for one minute and 72 °C, 30 seconds; finally at 72 ºC for 7 min, using a Nyx Technik® Thermocycler. For the PCR-RFLP 50 µg of PCR product was used which was digested with 1.5 U of Bg/II in a final volume of 25 µL. It was incubated for 16 hrs and the digestion products were separated by electrophoresis in a 1 % agarose gel in 1X TBE solution; a standard 50 bp marker was used. The gels were stained with ethidium bromide and the images were captured with the DigiDoc-IT System® photodocumenter and the Launch Doc-ITLS Acquisition® software. A pattern of two fragments (194 and 291 bp) corresponding to the European mythotype (E) and a single undigested fragment of 485 bp corresponding to the African mitotype was visualized (Crozier and Crozier, 1993).
For the detection of the L2S1int polymorphic site, the primers F: GGCGTCCAGGTAACCGTCTCC and R: CGGTTGGAGGCGAACGGAAA were designed to obtain an 830 bp fragment. The PCR conditions are those recommended by Suazo and Hall (2002), using the restriction enzyme AVAI to identify the African allele. Since this fragment analysis were inconclusive to identify the expected molecular characteristics, the results are not included in this manuscript.
For the statistical analysis and to determine the Hardy-Weinberg equilibrium, the heterozygous deficit and the polymorphic content index of the locus used for the characterization of the populations, the Cervus v3.03® and GENEPOP v4.0.10® softwares were used.
RESULTS
The molecular method used in the present work was based on the PCR-RFLP of DNAn and DNAm; On the one hand, the 830 bp L2S1int suballele nuclear fragment was amplified using the restriction enzyme AvaI. Likewise, the mitochondrial fragment of 485 bp of the mitochondrial gene of cytochrome b was amplified using the enzyme Bg/II; since these two loci contain the changes in the sequences that allow discrimination of the African subspecies Apis mellifera scutellata and the non-African subspecies.
Figure 1 shows the agarose gels with the mRNA PCR products, electrophoresed and stained with ethidium bromide (EtBr). The amplified fragment of the cytochrome b gene with a band size of 485 bp (A) and the amplified segment of the suballele L2S1int a fragment of 830 bp (B) was observed. In the present work it was observed that the polymorphism of the mitochondrial gene of Cytochrome b detected with the enzyme Bg/II discriminates the mitochondrial haplotype of A. m. scutellata; ancestor of Africanized bees, from the haplotype of A. m. mellifera, A. m. ligustica, A. m. cárnica and A. m. caucasica (figure 2).
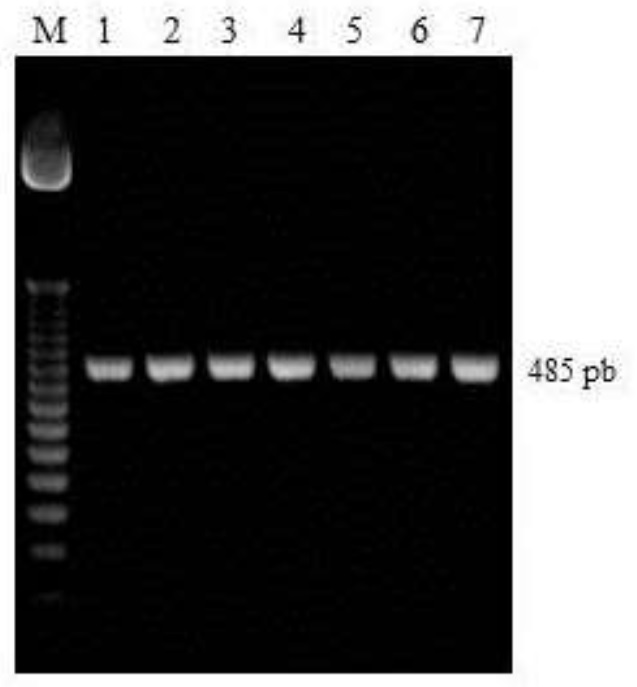
Lanes: 1, 50 bp molecular marker and Lanes 2-8, segment of the amplified cytochrome b gene (A).
Figura 1 1.7% agarose gel electrophoresis with the 485 bp amplified fragment of the cytochrome b gene.
The molecular method used in the present work was based on the PCR-RFLP of DNAn and DNAm; on the one hand, the L2S1int suballele nuclear fragment of 830 bp was amplified using the restriction enzyme AvaI. Likewise, the mitochondrial fragment of 485 bp of the mitochondrial gene of cytochrome b was amplified using the enzyme Bg/II; since these two loci contain the changes in the sequences that allow discrimination of the African subspecies Apis mellifera scutellata and the non-African subspecies.
Figure 1 shows the agarose gels with the mRNA PCR products, electrophoresed and stained with ethidium bromide (EtBr). The amplified fragment of the cytochrome b gene with a band size of 485 bp (A) and the amplified segment of the suballele L2S1int a fragment of 830 bp (B) was observed.
In the present work it was observed that the polymorphism of the mitochondrial gene of Cytochrome b detected with the enzyme Bg/II, discriminates the mitochondrial haplotype of A. m. scutellata; ancestor of Africanized bees, from the haplotype of A. m. mellifera, A. m. ligustica, A. m. cárnica and A. m. caucasica (figure 2).
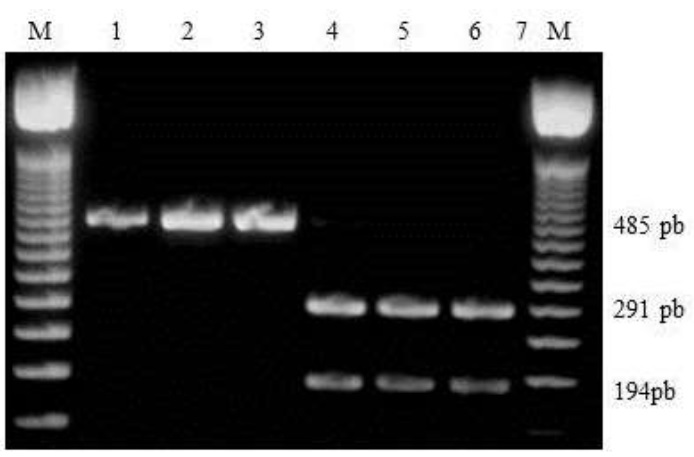
1.7% agarose gel electrophoresis. Fragment of 485 bp characteristic of Africanized bees (lanes 2-4). Segments of 194 Fragment 291 bp characteristic of non-Africanized bees (lanes 5-7).
Figura 2 RFLP patterns of the cytochrome b / Bg / II gene segment.
Electrophoretic analysis of 1.7 % agarose gels with the DNAm of the samples showed fragments of DNAm bands of African and European genotypes (Figure 3). The fragments that maintained the size of 485 bp are characteristic of African bees, (because they lack the cutting site for the enzyme Bg/II in the cytochrome b gene); while in individuals that had the restriction site fragments of 194 and 291 bp were produced, corresponding to non-African bees.

Segments of 485 bp characteristic of Africanized bees (lanes 2-13, 15 and 18). Segments of 194 and 291 bp characteristic of European bees (lanes 14, 16, 17).
Figura 3 RFLP patterns of the cytochrome b gene segment obtained with the Bg/II endonuclease. 1.7 % agarose gel electrophoresis.
It was observed that the populations have a degree of Africanization greater than 55 % (Table 1), with the exception of Jaumave.
Table 1 Number and frequencies (percentage) of the distribution of European and African DNAm patterns in Tamaulipas bee populations
| Locality | N | European (E) | African (A) | E-A |
| Victoria | 15 | 5(33.3) | 9(60.0) | 1(6.7) |
| Altamira | 24 | 2( 8.3) | 22(91.7) | |
| Llera | 23 | 5(21.7) | 18(78.3) | |
| Güémez | 24 | 8(33.3) | 14(58.3) | 2(8.3) |
| Padilla | 12 | 5(41.7) | 7(58.3) | |
| Jaumave | 5 | 2(40.0) | 2(40.0) | 1(20.0) |
| Total | 103 | 27(26.2) | 72(69.9) | 4(3.9) |
On the other hand, genotypic and allelic frequencies in bee populations in the Tamaulipas regions are presented in Table 2.
Table 2 Genotypic and allelic frequencies in Tamaulipas bee populations
| Population | N | Genotypic Frequencies | ||
| European (E) | African (A) | E/A | ||
| Victoria | 15 | 0.333 | 0.600 | 0.067 |
| Altamira | 24 | 0.083 | 0.917 | |
| Llera | 23 | 0.217 | 0.780 | |
| Güémez | 24 | 0.333 | 0.583 | 0.083 |
| Padilla | 12 | 0.417 | 0.583 | |
| Jaumave | 5 | 0.400 | 0.400 | 0.100 |
E/A = European / African
The performed analyzes for the genetic frequencies of the populations indicated that they are not in Hardy-Weinberg equilibrium, probably because there is a heterozygous deficit.
DISCUSSION
The molecular method used allowed discrimination of the African subspecies Apis mellifera scutellata and non-African subspecies (Zamora et al., 2008 Esquivel-Rojas et al., 2015). The polymorphism of the mitochondrial gene of cytochrome b detected with the Bg/II enzyme discriminates the mitochondrial haplotype of A. m. scutellata; ancestor of Africanized bees, from the haplotype of A. m. mellifera, A. m. ligustica, A. m. cárnica and A. m. caucasica (Pinto et al., 2003 Genchi-García et al., 2018).
Electrophoretic analysis of 1.7 % agarose gels with the DNAm of the samples showed fragments of DNA bands of African and European genotypes. The fragments that maintained the size of 485 bp are characteristic of African bees, (because they lack the cutting site for the Bg/II enzyme in the cytochrome b gene); while in individuals who had the restriction site, fragments of 194 and 291 bp were produced, corresponding to non- African bees.
The pattern observed in this work coincides with the pattern to discriminate African haplotypes from non-Africans in Africanized areas (Pinto et al., 2003 Tibatá et al., 2018). It was observed that the populations have a degree of Africanization; similar results were reported by Esquivel-Rojas et al. (2015), Medina-Flores et al. (2015), Quezada-Euán (2007) and Zamora et al. (2008), when evaluating the polymorphism of Africanized populations in Mexico.
The results obtained with the modifications to the DNAm purification protocol, allowed to know the genetic structure of honey bees in the center of Tamaulipas. The characterization of the populations showed that 69.9 % of the colonies evaluated had in their DNAm, the restriction site for the African haplotype; as noted in the literature (Quezada-Euán, 2007; Zamora et al., 2008 Esquivel-Rojas et al., 2015; Medina-Flores et al., 2015). While 26.2 % of the sampled apiaries had the haplotype in their DNAm, which characterizes them as of European maternal origin. 4 % of the samples presented mixed European-African haplotype; this could be due to drones being wild and presenting a high proportion of African genes. However, Tamaulipas beekeepers have as strategies to introduce queens every year or year and a half, but certified before the Ministry of Agriculture, Livestock, Rural Development, Fisheries and Food (SAGARPA), if they are not carriers of the African gene (SAGARPA, 2016 ).
In the sampled sites, the African haplotype represented a high percentage of the colonies, confirming the reduction of European type alleles in the beekeeping populations of Mexico, Colombia and Brazil (Quezada-Euán, 2007); and the dominance of African nuclear and mitochondrial genes in the genome of domestic colonies, from the arrival of the Africanized bee in Mexico.
Altamira populations are composed almost entirely of bees with mitotypes of African origin, probably because the producers do not make the annual change with queens of European origin. These results suggest that beekeeping practices, such as the annual change of queens, have not been sufficient to reverse the frequency of African genes in local populations. In addition, the tropical climatic conditions of the region have facilitated the displacement of European genes.
It was observed that populations have a high degree of Africanization, with the exception of Jaumave; which indicates that it is still necessary to continue introducing European certified queens (SAGARPA, 2016).
Quezada-Euán (2007) reported 95.0% of Africanization for Chiapas and Tabasco; Uribe et al. (2003) and Zamora et al. (2008) reported 13.7, 48.0, 50.0 and 21.0 %, of haplotypes derived from African bees in Sonora, Baja California Sur and Baja California Norte, respectively. Similarly, Quezada-Euán (2007) reported that the proportion of Africanization in Michoacán and Jalisco was 56.0 and 40.0 %, respectively. While in Yucatan, it increased from 61.0 to 73.0 %. Alaniz-Gutiérrez et al. (2016) observed that African morphotypes in Ensenada and Mexicali, Baja California are above 85.0 %.
CONCLUSION
It can be concluded that the populations of Apis mellifera L. de Tamaulipas, are constituted by characteristic genes A. mellifera scutellata and to a lesser extent by genes of European subspecies such as A. mellifera ligustica. The results of the molecular marker analysis showed that Tamaulipas populations are heterogeneous with introgression of African genes in European populations.
ACKNOWLEDGMENT
The authors thank the beekeepers in the downtown area of Tamaulipas. To the Laboratory of the Faculty of Veterinary Medicine and Zootechnics-UAT. To the Council of the National System of Technological Education (CoSNET) for partial financing for the development of the project.
REFERENCES
ALANIZ-GUTIÉRREZ L, Torres-Salado N, Ail-Catzim CE, Velazco-López JL. 2016. Frecuencia de morfotipos africanizados y europeos de Apis mellifera en Ensenada y Mexicali, Baja California. Ecosistemas y recursos agropecuarios. 3(9):421-426. ISSN: 2007-9028. [ Links ]
CASTAÑEDA-VENEGAS JÁ. 2011. Producción de miel de abejas de flor de azahar. Instituto Interamericano de Cooperación para la Agricultura (IICA). México, D.F. [ Links ]
CLARKE KE, Oldroyd BP, Quezada EJJG, Rinderer TE. 2001. Origin of honeybees (Apis mellifera L) from the Yucatan peninsula inferred from mitochondrial DNA analysis. Molecular Ecology. 10:1347-1355. DOI:10.1046/j.1365-294X.2001.01274.x [ Links ]
CROZIER RH, Crozier Y. 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 133(1):97-177. ISSN: 1943-2631. [ Links ]
ESQUIVEL-ROJAS S, Macías-Macías JO, Tapia-González JM, Contreras-Escareño F, León-Mantecón MJ, Silva-Contreras A. 2015. Selección de abejas (Apis mellifera L) con baja defensividad y su relación con el ambiente en Jalisco, México. Abanico Veterinario. 5(1):44-50. ISSN: 2007-428X. [ Links ]
GENCHI-GARCÍA ML, Reynaldi FJ, Bravi CM. 2018. An update of Africanization in honey bee (Apis mellifera) populations in Buenos Aires, Argentina. Journal of Apicultural Research. 57(5):611-614. https://doi.org/10.1080/00218839.2018.1494887. [ Links ]
GUZMÁN-NOVOA E, Hunt GJ, Uribe-Rubio JL, Prieto-Merlos D. 2004. Genotypic effects of honey bee (Apis mellifera) defensive behaviour at the individual and colony levels: the relationship guarding, puirsing and stinging. Apidologie. 35(1):15-24. ISSN: 0044-8435, DOI: 10.1051/apido:2003061 [ Links ]
MEDINA-FLORES CA, Guzmán-Novoa E, Hamiduzzaman MM, Aguilera-Soto J, López- Carlos MA. 2015. Africanización de colonias de abejas melíferas (Apis mellifera) en tres regiones climáticas del norte de México. Veterinaria México OA. 2(4):1-9. ISSN: ISSN 2448-6760. [ Links ]
PINTO MA, Spencer JJ, Rubink WL, Coulson RN, Patton JC, Sheppard WS. 2003. Identification of Africanized Honey Bee (Hymenoptera: Apidae) mitochondrial DNA: Validation of a Rapid Polymerase Chain Reaction-Based Assay. Annals of the Entomological Society of America. 96:679-684. ISSN: 0013-8746. [ Links ]
QUEZADA-EUÁN JJG. 2007. A retrospective history of the expansion of Africanized honeybees in Mexico. Journal of Apicultural Research and Bee World. 46(4):295-300. ISSN: 0021-8839. doi.org/10.1080/00218839.2007.11101412 [ Links ]
SAGARPA (Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación). 2007. Situación actual del pequeño escarabajo de la colmena en México y sus perspectivas. 17ª. Reunión Anual de la CONASA. [online]. Disponible: http://www.conasamexico.org.mx/conasa/docs_17a_reunion/comite07/Igor_Romero_So sa.pdf [citado 12 de diciembre de 2016]. [ Links ]
SAGARPA (Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación). Reglas de Operación de los Programas de la Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación para el ejercicio fiscal 2016. [online]. Disponible: http://www.gob.mx/cms/uploads/attachment/file/44530/Reglas-Operacion-2016-sagarpa.pdf [citado 12 de diciembre de 2016]. [ Links ]
SDR (Secretaría de Desarrollo Rural). 2017. Tamaulipas segundo lugar a nivel nacional en producción de cítricos. [online]. Disponible: Disponible: https://www.tamaulipas.gob.mx/desarrollorural/2017/04/tamaulipas-segundo-lugar-a- nivel-nacional-en-produccion-de-citricos/ [citado 13 de septiembre de 2019]. [ Links ]
SIFUENTES RAM, Puentes-Montiel HG, Moreno-Medina VR, De La Rosa-Reyna XF. 2006. Assesment of the myostatin Q204X allele using an allelic discrimination assay. Journal of Genetic and Molecular Biology. 29(3):496-497. ISSN: 1415-4757, doi.org/10.1590/S1415-47572006000300017 [ Links ]
SUAZO A, Hall HG. 2002. Nuclear DNA PCR-RFLPs that distinguish African and European honeybee groups of subspecies. II: Conversion of long PCR markers to standard PCR. Biochemical Genetics. 40(7/8):241-261. ISSN: 0006-2928. [ Links ]
TIBATÁ VM, Arias E, Corona M, Ariza-Botero F, Figueroa-Ramírez J y Junca H. 2018. Determination of the Africanized mitotypes in populations of honey bees (Apis mellifera L.) of Colombia. Journal of Apicultural Research . 57(2):219-227. https://doi.org/10.1080/00218839.2017.1409065 [ Links ]
URBINA-ROMERO RA, Utrera-Quintana F, Castillo-González F, Livera-Muñoz M, Benítez-Riquelme I, Villa-Mancera AE, Hernández-Hernández JE, Silva-Rojas HV. 2019. Valoración del origen africanizado en la integración de una población experimental de Apis mellifera L. Revista fitotecnia mexicana. 42(2):111-118. http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S0187-73802019000200111&lng=es&tlng=pt [ Links ]
URIBE RJL, Guzmán-Novoa E, Hunt GJ, Correa BA, Zozaya J. 2003. Efecto de la africanización sobre la producción de miel y comportamiento defensivo y tamaño de las abejas melíferas (Apis mellifera L.) en el Altiplano Mexicano. Veterinaria México. 34(1):47-59. ISSN: 0301-5092. [ Links ]
VÁSQUEZ-CASTRO JA, Narrea-Cango M, Bracho-Pérez JC. 2006. Efecto del ácido oxálico, ácido fórmico y coumaphos sobre Varroa destructor (Acari: Varroidae) en colonias de abejas. Revista Peruana de Entomología. 45:149-152. ISSN: 2222-2529. [ Links ]
VILLEGAS DG, Bolaños MA, Miranda SJ, García AJ, Galván GO. 2016. Flora nectarífera y polinífera en el Estado de Tamaulipas. SAGARPA. [online]. Disponible: Disponible: http://www.agrotamaulipas.gob.mx/informacion_sector/forestal/Flora%20Tamaulipas.pdf [citado 12 de diciembre de 2016]. [ Links ]
ZAMORA O, Domínguez R, Alaniz-Gutiérrez L, Quezada-Euán JG. 2008. Frequency of European and African-derived morphotypes and haplotypes in colonies of honey bees (A. mellifera) from NW México. Apidologie. 39:388-396. ISSN: 0044-8435, DOI: 10.1051/apido:200801. [ Links ]
Received: June 20, 2019; Accepted: October 05, 2019; Published: November 05, 2019











 texto en
texto en 



