INTRODUCTION
The apple tree (Malus domestica Borkh) is an important temperate-climate fruit tree in Mexico, with its 2015 production totaling 750 324.85 t. It is grown mainly in the states of Chihuahua, Durango, Puebla and Coahuila; in the last state, the main production is in the Sierra de Arteaga, with an annual production of 26 211.18 t of apple in 5 770 ha (SIAP 2016), in which the Red Delicious and Golden Delicious cultivars are planted (Contreras de la Reé et al. 2010). Agricultural production based on a few cultivars is subject to risks, because genetic uniformity is vulnerable to pests, diseases or environmental stress (Granados et al. 2009), as occurred with the devastation of the potato crop by Phytophthora infestans in Ireland (Woodham- Smith 1991), or in the case of the current problem facing U.S. apple (Malus) production due to the low number of varieties cultivated (Gayle et al. 2015). One way to avoid problems due to pests or diseases is to maintain genetic diversity in the crops, through the conservation of different cultivars (Granados et al. 2009). The greater the genetic diversity in a breeding program, the better the chances of developing new cultivars with resistance to diseases, quality and productivity (Smolik and Krzysztoszek 2010).
The effectiveness of breeding programs depends on the nature and magnitude of the variability (Lado et al. 2012), and on the use of molecular genetic markers, which allow specifying and quantifying the phylogenetic relationships between closely related species (Savelyeva and Kudryavtse 2015). Within the genetic markers are the DNA markers, such as the Inter-Simple Sequence Repeats (ISSR), which besides being highly polymorphic allow obtaining a variation in DNA regions between the microsatellites that are dispersed throughout the genome (Rizkalla et al. 2012). ISSRs have been used to identify genotypes in crops such as pear (Kalkisim et al. 2016), mango (Singh et al. 2007), mulberry (Prasanta et al. 2008), papaya (Carrasco et al. 2009) cucumber and melon (Parvathaneni et al. 2011), wheat (Rizkalla et al. 2012), and birch (Pan et al. 2006), among others. These markers have been shown to have an advantage for apple diversity studies (Smolik and Krzysztoszek 2010, He et al. 2011). Based on the above and in view of the little genetic information available about the apple cultivars established in the Sierra de Arteaga, Coahuila, the objective of this study was to determine the genetic diversity of 12 apple cultivars adapted to the climatic conditions of the Sierra de Arteaga, Coahuila through microsatellites (ISSR).
MATERIALS AND METHODS
Plant material
Leaves were collected from 12 apple cultivars whose chilling requirements vary between 400 and 600 chilling units (CU). The cultivars were: Corail, Imperial Gala, Honey Crisp, Gale Gala, Glory, Buckeye Gala, Cameo, Pink Lady, Española, Royal Gala and Pacific Gala. The cultivar Golden Delicious was used as a control; it has requirements between 900 and 1 200 CU. Plant tissue sampling was carried out in the town of San Antonio de las Alazanas, Municipality of Arteaga, Coahuila.
DNA Extraction
The extraction of DNA was done from healthy leaves of each cultivar, with the modified CTAB technique. From each leaf, 0.1 g was taken and ground with liquid nitrogen. 1X CTAB lysis solution [5 M NaCl, 0.5 M EDTA (pH 8), 1 M Tris 1 M (pH 8) and 0.5% 2-mercaptoethanol] was used. The obtained material was washed with chloroform-isoamyl alcohol (24:1 V/V) and the supernatants were washed with 99% ethanol, after which they were left for 60 min at -20 °C. After this, the sample was centrifuged at 13 300 g for 15 min and the supernatant was discarded, the formed pellet was washed three times with 70% ethanol, left to dry at room temperature and stored in 50 µL of TE (1X) plus 10 µL of RNase prepared at a concentration of 1 µg µL-1. DNA integrity was tested on a 1% agarose gel and quantification was performed with the Gen5 1.11 DNA program.
ISSR Analysis
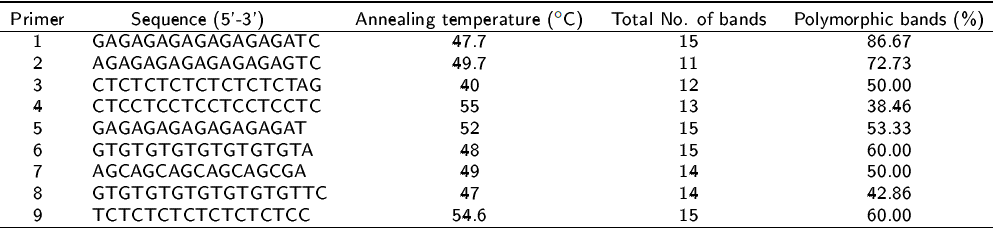
Nine primers (Table 1) were tested at different temperatures in standard condition and then amplification conditions were optimized by gradient PCR, in order to obtain clear bands. Amplification was performed with polymerase chain reaction (PCR) in a volume of 11.5 µL, composed of 3.0 µL of ddH2O, 2.17X of 10X buffer solution added with MgCl2, 0.21 mM of the dNTP’s mixture, 2.6 µM of each primer, 8.7% of DMSO, 0.1 U of PaqDNA polymerase at a concentration of 5 U µL-1 and DNA. In a Px2 Thermal Cycler, PCR was performed with an initial denaturation temperature of 1 min at 94 °C, 40 cycles composed of denaturation of 1 min at 94 °C, annealing of 45 s and extension of 2 min at 72 °C, followed by an extension step of 7 min at 72 ºC. The amplification products were visualized on 1.5% agarose gels with SB 1X buffer and stained with ethidium bromide. With an Axygen Biosciences 100 bp marker, the size of each of the fragments obtained was determined.
Statistical Analysis
Each band obtained in the agarose gels was considered as a locus and codified as absence (zero) and presence (one) for each cultivar studied. All analyses were performed with the InfoGen v. 2011 program (Di Rienzo et al. 2008) to calculate the percentage of polymorphic ISSR, genetic diversity, Nei’s unbiased heterozygosity and effective number of alleles. In order to cluster the different genotypes evaluated, a dendrogram was created using Sokal Sneath distance 3, since it was the one with the highest cophenetic correlation (0.888).
RESULTS AND DISCUSSION
The molecular characterization of the 12 apple samples was carried out with the ISSR technique. The advantage of this molecular tool lies in its great capacity to detect differences between individuals that may be related (Azofeifa-Delgado 2006). Using nine ISSR primers, we amplified a total of 124 bands, of which 63.71% had polymorphism. An example of the DNA polymorphism patterns generated by the first primer [(GA) 8T] is shown in Figure 1. Results indicate a diversity of 0.24 and average number of alleles of 1.637, indicating genetic variability. Some authors point out that ISSR markers allow finding high levels of polymorphism between individuals and are widely used for the identification of cultivars of the Malus species. In this regard, Calvo et al. (2014) determined the genetic variability of 23 populations of wild apple trees in the Andean mountain range, finding a total of 46 alleles with 10 SSR primers and a genetic variation of 66% among the populations. In the same vein, He et al. (2011) evaluated wild apple trees with 20 ISSR primers and report 110 polymorphic bands and a similarity coefficient of 0.74 to 0.94 among cultivars. On the other hand, Smolik and Krzysztoszek (2010) evaluated the genetic variability of eight apple tree cultivars, among which Golden Delicious stands out, confirming the great advantage that the ISSR have against other traditional methods of characterization and estimation of diversity, since this type of molecular marker is not affected by environmental conditions.

Figura 1: DNA amplification patterns generated by the primer (GA)8T in the samples, lane M= molecular marker, 1= Corail, 2=Imperial Gala, 3=Honey Crisp, 4=Gale Gala, 5=Glory, 6=Buckeye Gala, 7=Cameo, 8=Pink lady, 9=Española, 10=Royal Gala, 11=Pacific Gala, 12=Golden Delicious.
The number of cultivars evaluated in the present study is small, but with the bands found, a dendrogram (Figure 2) was obtained that shows the formation of two groups. Group one is formed by the cultivars Gale gala, Royal gala and Pacific gala, with chilling requirements that oscillate around 400 CU. In addition, these three cultivars present similar banding patterns and extra bands at 250 bp with the primers six, eight and nine; this same group includes the Golden Delicious control sample, which despite requiring between 900 and 1 200 CU to leave dormancy and start to flower (Contreras de la Reé et al. 2010) shares band similarity with Royal gala in most of the patterns generated with the primers evaluated.
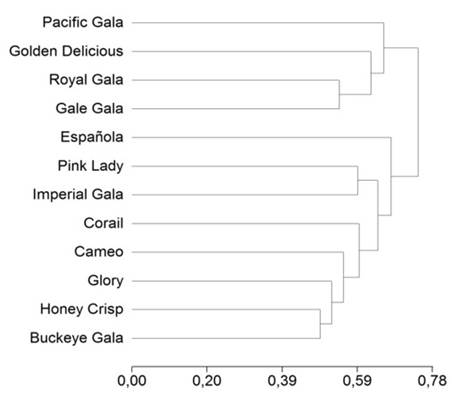
Figura 2: Clustering of 12 commercial apple tree cultivars, grown in the Arteaga region, Coahuila, based on ISSR markers with the Sokal Sneath 3 distance matrix.
Group two includes eight cultivars, the Spanish cultivar being the most different. Within this group are the cultivars Buckeye gala and Imperial gala, which require between 450 and 500 CU to flower; in addition, the fruit that these two cultivars produce is redder and its color is established up to 10 d before Pacific gala and Royal gala, while Buckeye gala is one of the first cultivars to be harvested (Contreras de la Reé et al. 2010). The two cultivars that showed a high level of similarity between them were Honey crisp and Buckeye gala, with a distance of 0.49, despite presenting different chilling requirements. These two cultivars have the same banding pattern with primers three and four, in addition to minimal banding differences with the other seven primers between 100 and 400 bp. These two cultivars are joined by Cameo, which is the only one that presents a band at 500 bp with primer number eight and at 550 bp with primer number 6.
Results indicate a good molecular characterization in the cultivars evaluated, but the genetic diversity found does not correlate with the fruit quality, chilling requirement or flowering time that the cultivars present. The level of polymorphism found among them suggests proposing the establishment of plantations with a greater diversity of cultivars, adapted to the climatic conditions of the region, so as not to depend on only a few cultivars for apple production in the Sierra de Arteaga, Coahuila.











 nueva página del texto (beta)
nueva página del texto (beta)