INTRODUCTION
Foodborne illnesses (FBIs) are a syndrome caused by the ingestion of foods containing etiological agents in quantities that affect the consumer’s health (Soto et al. 2016). The FBIs is related to the physical damage that they cause, and these are associated with negative socio-economic impact, personal afflictions and whealth services (Zotta 2015). As a result of the growing demand for the production, marketing and consumption of food, governments require a more efficient and strict control of their hygienic and sanitary quality, in order to prevent diseases (Martino et al. 2010). FBIs constitute a national and global public health problem, due to the increase in their occurrence, the emergence of new forms of transmission, the appearance of vulnerable population groups, the increased resistance of pathogens to antimicrobial compounds and the socioeconomic impact that they cause (Rodríguez et al. 2015). One of the main sources of contagion of pathogenic bacteria or poisoning is the consumption of contaminated food, including fish, seafood, meat products, poultry, dairy products, vegetables, fresh eggs and even honey (Rojas and González 2006, FAO 2009, López et al. 2013).
Listeriosis is a foodborne disease caused by Listeria monocytogenes and its importance is due to its clinical impact, high mortality rate and the economic effect resulting from outbreaks associated with food consumption (Castañeda et al. 2014). According to Orihuel et al. (2011), the importance of food as the primary transmission route of listeriosis to people was not recognized until the 1980s, following several major disease outbreaks in North America and Europe. An important characteristic of this foodborne disease is that the pathogen can multiply at refrigeration temperatures between 4 and 5 °C, until reaching significant figures. Although there are many and diverse foods that can be contaminated with L. monocytogenes, outbreaks and sporadic cases of listeriosis are predominantly associated with ready-to-eat foods. In Mexico, few studies have been conducted for the detection of L. monocytogenes in foods (Castañeda et al. 2014). The government agency responsible for maintaining these records in Mexico is the National Epidemiological Surveillance System that reports that in 2016 there were 40 246 cases of paratyphoid fever and salmonellosis, 17 644 cases of typhoid fever, and 2 021 298 cases due to intestinal infections caused by different organisms (SINAVE 2016).
For several decades, infections by enteropathogens have been diagnosed through the cultivation of food samples and the bacteria growing in the culture media have been identified using morphological and physiological criteria, with the disadvantage of not being able to obtain results for five days or more, depending on the type of pathogen to be identified (Scheu et al. 2009). This makes it necessary to develop fast and sensitive techniques that allow pathogens to be detected in raw or processed foods, such as Polymerase Chain Reaction or PCR (Rojas and González 2006). Therefore, the objective of the work was to standardize the PCR technique for the diagnosis of Listeria monocytogenes in chicken, beef and pork.
MATERIALS AND METHODS
The research was carried out in the Animal Nutrition Laboratory of the Colegio de Postgraduados, Campus Montecillo, belonging to the Postgraduate Program in Genetic Resources and Livestock Productivity, located in Montecillo, Texcoco de Mora, State of Mexico.
The standardization of the Polymerase Chain Reaction (PCR) technique for the diagnosis of L. monocytogenes in chicken, beef and pork was carried out based on the microbiological method approved in the Official Mexican Standard NOM-143-SSA1-1995. For the detection of this bacterium by PCR, 20 samples of 250 g were collected for the three meats. Samples were collected randomly at different points of sale in the city of Texcoco, State of Mexico. The sampling was repeated three times at each point of sale at 15 d intervals. The sample size was calculated by means of stratified sampling (Castillo 2005). The meat samples were collected at 9:00 a.m. and transferred to the laboratory for analysis at 4 °C, under aseptic conditions according to the specifications of standard NOM-109-SSA1-1994.
The strains used in this study were L. monocytogenes (ATCC 19115) as a positive control, while Escherichia coli ATCC 25922 was used as a negative control. The strains were preserved in trypticas soya broth (TSB) with 15% glycerol at -20 °C in a Thermo Scientific® model 88500A61 ultra-freezer.
Artificial inoculation
The meat samples used for the inoculation were confirmed as negative by microbiological analysis and by PCR for L. monocytogenes. Inoculation of the meat was performed with 2.1 x 102 cfu mL-1 of a culture of L. monocytogenes and E. coli incubated for 48 h in TSB at 30 °C. To validate the detection limits of L. monocytogenes, each dilution had an enrichment phase in accordance with the provisions of Official Mexican Standard NOM-143-SSA1-1995. At the end of enrichment, decimal dilutions from 10-1 to 10-10 were made to determine the minimum detection limit of CFU by PCR. Dilutions were incubated on OXA Agar plates at 37 °C for 24 h and the correct ratio between the number of CFUs and their corresponding dilutions was measured at 2.1 x 102 cfu g-1 for L. monocytogenes.
Detection of Listeria monocytogenes by the microbiological method
The detection of L. monocytogenes was carried out as indicated in the Official Mexican Standard NOM-143-SSA1-1995 using the Oxford (OXA) and trypticase soy agar with yeast extract (TSAYE) media, incubated at 35 °C for 24 and 48 h. After incubation, the suspected colonies were subjected to the following biochemical tests: Gram-staining, hemolysis (5% sheep blood agar), mobility in agar (SIM medium), and the Cristie-Atkins-Munch-Peterson (CAMP). Additionally, the biochemical tests established by the protocol of the Micro-ID® Listeria Identification System were carried out.
Detection of Listeria monocytogenes by PCR
DNA was extracted from the chicken, beef and pork samples using the commercial Wizard® Genomic DNA Purification kit. For the detection of the genus Listeria, prs gene amplification assays were carried out by PCR, using the primers PRS F 1 and PRS R (5’-GCTGAAGAGATTGCGAAAGAAG-3’ and 5’-CAAAGAAACCTTGGATTTGCGG-3’) that amplify a 370-bp fragment of the prs gene (Callejo et al. 2008, Doumith et al. 2004), and for Listeria monocytogenes, the primers MAR 1 - MAR 2 (5’-GGGCTTTATCCATAAAATA-3’ and 5’-TTGGAAGAACCTTGATTA-3’) that generate a 453-bp fragment of the iap gene (Cocolin and Rantsiou 2016, Lozano et al. 2013). DNA from L. monocytogenes ATCC 19115 was used as a positive control, and DNA from E. coli ATCC 25922 as a negative control. Additionally, a DNA-free tube was used as the blank for system control.
The PCR was performed with an initial denaturation cycle of 95 °C for 1 min, 35 cycles at 94 °C for 30 s of denaturation, 20 s at 53 °C of hybridization-extension and a final extension cycle of 30 s at 74 °C; at the end an extension of 8 min was made at 74 °C. The program was implemented in a BIO RAD Model c1000 Touch Thermal Cycler. For all tests, the amplified products were evaluated on a 2% agarose gel, using the 100-bp molecular weight ladder marker. The minimum DNA concentration detectable with the PCR technique was done by inoculating meat samples with different inocula of L. monocytogenes. The DNA concentration ( mL-1) was determined by using a Nanodrop Model 2000c spectrophotometer and the concentration of bacteria (CFU mL L-1) by plate count.
The specificity of the primers that amplify the prs gene in Listeria and the iap gene in L. monocytogenes was evaluated by PCR with E. coli DNA to rule out nonspecific amplifications. The calculation of sensitivity (S) and specificity (Sp) was made using the McNemar test and the Kappa index (López and Fernández 2001). To determine the concordance between the microbiological method and the PCR technique in the detection of L. monocytogenes, the Kappa index was used (López and Fernández 2001, Aguilera 2005), which was calculated as k = (Po-Pe)/(1-Pe), where Po= observed agreement and Pe= expected agreement. The interpretation of the Kappa index was made with the following scale: 0 to 0.2, insignificant; 0.21 to 0.4, medium; 0.41 to 0.6, moderate; 0.61 to 0.8, substantial; 0.81 to 1 almost perfect (Redondo 2015). In the case of the McNemar tests, a significance level of α= 5% was established.
In order to estimate the level of concordance between the PCR and microbiological methods, it was decided to estimate the Kappa index (López and Fernández 2001), because it reduces the possible concordance between the two methods due to chance; on the other hand, to calculate the conditional probability of detection by the PCR method given a positive or negative result of the microbiological one, Bayes’ Theorem was used (Díaz and de la Fuente 2006, López et al. 2007).
RESULTS
Analytical sensitivity of PCR
The assay to determine the minimum DNA concentration was performed by amplifying the prs gene for Listeria (370 bp) and the iap one for L. monocytogenes (453 bp) (Figure 1). The decreased intensity of the signal resulting from the amplifications of the prs (Listeria) and iap (L. monocytogenes) genes is presented from lane 7 (10-5 dilution) in which no signal was recorded, indicating that the minimum concentration detectable by the developed PCR is in the 10-4 dilution (3 ng μL-1) in lane 6.

Figura 1: Minimum DNA detection by PCR for the iap (Listeria monocytogenes) and prs (Listeria spp) genes by 2% agarose gel electrophoresis. Lane: 1) 100-bp ladder marker; 2) negative control; 3) 10-1 dilution, 4) 10-2 dilution; 5) 10-3 dilution; 6) 10-4 dilution, 7) 10-5 dilution, 8) 100-bp ladder marker.
Relative specificity
As a result of the amplification of the iap and prs genes of the inoculated samples and without inoculating with L. monocytogenes (ATCC 19115), 100% sensitivity and specificity were obtained in both cases (Figure 2). It was observed that the samples intentionally contaminated with L. monocytogenes amplified the region of the mentioned genes; on the other hand, in Figure 3 it can be seen that the samples contaminated with E. coli (ATCC 35922) did not present a signal. For this reason, the primers MAR 1 - MAR 2 and PRS F-PRS R were highly specific for the amplification of the iap and prs genes, respectively (Table 1).
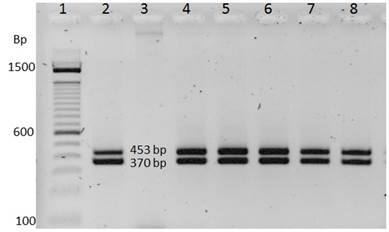
Figura 2: Amplification of the iap (453 bp) and prs (370 bp) genes for samples positive for Listeria monocytogenes. Lane 1: 100-bp DNA ladder marker, Lane 2: positive control, Lane 3: negative control for E. coli (ATCC 35922), Lane 4-8: Listeria monocytogenes DNA (ATCC 19115).

Figura 3: Amplification of the iap (453 bp) and prs (370 bp) genes for samples negative for Listeria monocytogenes. Lane 1: 100-bp DNA ladder marker, Lane 2: positive control of Listeria monocytogenes (ATCC 19115), Lane 3: negative control, Lane 4-8: samples inoculated with the pure strain of E. coli (ATCC 35922).
Comparison of the microbiological method and PCR
The comparison of the PCR and microbiological methods for the detection of L. monocytogenes in chicken, beef and pork samples is presented in Table 2. To evaluate the PCR results, McNemar’s analysis was used to identify possible differences between the evaluated methodologies; and second to estimate the degree of concordance between these. Differences (p > 0.05) were found between the methods and the chicken, beef and pork samples, as there was an 89.4% level of agreement between the values found (Table 2). According to the k values, an almost perfect degree of agreement between the methods was obtained, indicating the similarity in the results obtained between the microbiological and PCR methods for the detection of L. monocytogenes. Therefore, the detection of PCR is calculated based on the detection of the microbiological method, by means of Bayes’ Theorem. The conditional probabilities of Bayes’ Theorem, according to the values in Table 2, showed that when the microbiological method detected positive samples of L. monocytogenes, the PCR technique also detected them, whereas when the microbiological method did not detect L. monocytogenes, the PCR technique detected 12.5% more cases (Table 3).
Tabla 2: index (k) for detecting Listeria monocytogenes in chicken, beef and pork samples (n=60).

a= Samples that were positive in the PCR and in the microbiological method. b= Samples that were positive in the PCR and negative in the microbiological method. c= Samples that were negative in the PCR and positive in the microbiological method. d= Samples that were negative in the PCR and in the microbiological method. n= total of samples analyzed.
Tabla 3: Conditional probability of a positive detection by PCR (PCR+) given a positive result of the microbiological method (M+) and of a PCR+ given a negative microbiological result (M-).

The results of the Kappa index obtained for the samples showed an almost perfect level of agreement with values of 80, 91 and 100% (Table 4), which indicates that the PCR test developed provides results similar to the microbiological method for detecting L. monocytogenes. However, the PCR technique is more efficient and accurate in detecting L. monocytogenes, since the microbiological method was not always able to detect it.
DISCUSSION
The primers MAR 1-MAR 2 and PRS F-PRS R allowed us to obtain a minimum detectable concentration of 3 ng μL-1 by means of the PCR technique, which was lower than that reported by López and Mejía (2012) who evaluated organic solvents or PBS plus Tween20 as DNA extraction methods for L. monocytogenes, establishing a minimum detectable DNA concentration of 0.44 and 0.53 μg μL-1, respectively, while Amagliani et al. (2007) report minimum detectable DNA concentrations for L. monocytogenes of 5.3, 2.3 and 1.9 μg μL-1 with the Qiagen, Diatheva and Gentra commercial extraction kits; however, the purity of the DNA analyzed with these methods was low (1.64, 1.36 and 1.25), which could be due to contamination with particles such as proteins, which have inhibitory effects on DNA polymerase. In this regard Liu (2008) mentions that the purity of the amplified DNA is affected by multiple substances that interfere in the amplification processes of the nucleic acids, such as phenolic compounds, glycogen, calcium ions, fat and other organic substances (Liu 2013).
The primers MAR 1-MAR 2 and PRS F-PRS R were highly specific for the amplification of the iap and prs genes, observing a specificity and sensitivity of 100%,which is explain by the fact that the iap gene encodes a polypeptide of 484 amino acids containing a signal sequence of 27 amino acids, which produce a polypeptide of 47.5 to 60 kDa, which is essential for the survival of the bacterium, called p60, which is a cysteine residue in the C-terminal part that is conserved in all the proteins of other Listeria species. The mechanism of pathogenicity in L. monocytogenes has been shown to be related to the p60 protein, encoded by the iap gene, in which the region encoding a central domain of the p60 protein contains a tandem repeat sequence of ACAAAT, related to threonine and asparagine (TN), which is used to determine the molecular variation between strains of L. monocytogenes (Fagundes et al. 2008).
On the other hand, the house-keeping genes prs and ldh flank the prs-prfA-plcA-hly-mpl-actA-plcB-orfX-orfZ-orf B-orfA-ldh cluster, which consists of the 9.6 kb PrfA, a cluster of virulence-regulating genes of Listeria 1, LIPI-1. The prs gene encodes the enzyme phosphoribosyl pyrophosphate synthetase (318 amino acids) present in all Listeria species and is used for determination of the genus (Liu, 2013), which explains the specificity found in this study.
The level of concordance determined between the methods evaluated to detect samples positive for L. monocytogenes (89.4%, Table 2) in chicken, beef and pork supports the use of PCR to diagnose the presence of the bacterium, consistent with the results observed in other studies where concordance values of 100 and 98.43% were obtained when comparing the traditional microbiological method or the Gold Standard method with the PCR technique for the detection of L. monocytogenes, which indicates high reliability to detect samples positive for this bacterium (Poutou et al. 2005, Burbano et al. 2011).Therefore, the PCR technique is highly effective when compared to the traditional microbiological method for the diagnosis of L. monocytogenes in other matrices, since it allows obtaining Kappa indexes of 1 (Ramírez et al. 2010). The difference between the Kappa index values observed in the types of meat analyzed in the present study is explained by the possible obtainment of false positive PCR results, derived from the detection of the genetic material of nonviable bacteria (AINIA 2014) that the microbiological method cannot detect, since it works with DNA extracted from cells regardless of the moment of extraction. However, in the initial enrichment stage of samples with EB broth, the extraction of DNA from non-viable cells is minimized by increasing the number of live L. monocytogenes. Additionally, the PCR technique allows for complementary analyses on the same sample portion by means of traditional isolation techniques in culture, so that the possibility of obtaining a false positive with respect to the reference method is nil. On the other hand, there are foods that can inhibit the PCR reaction and therefore give false negative results. According to Rodríguez and Crespo (1999), Bayes’ Theorem refers to those situations where once an event B has been produced, it is a question of calculating whether it is due to a cause A. In other words, applied to the comparison of the methodologies evaluated in this experiment, with this theorem we look for the probability that the PCR technique will find a positive value, given that the microbiological method finds a positive value, or on the contrary, we look for the probability that the PCR technique will detect a positive value, given that the conventional method detects a negative value.
The value found by Bayes’ Theorem (Table 3) indicates that when the microbiological method detects a positive sample of L. monocytogenes, the PCR technique will also detect it. However, when the conventional method does not detect the presence of the pathogen, the PCR technique detects it by 12.5% more.
The variation in the results of both techniques evaluated in this study was also discussed by the AINIA (2014), which mentions the presence of false positives and negatives resulting from dead cells. This agrees with what was established by Candrian (1995) since the cells could be damaged due to the sanitation products, storage and in general by the methodology for sampling. Although there are few studies in which the Bayes’ Theorem methodology is applied to food microbiology, it is a practical tool that allows us to have a better predictive value of the presence of pathogens.
CONCLUSION
The primers MAR 1-MAR 2 and PRS F-PRS R were specific for the amplification of the iap and prs genes of L. monocytogenes and Listeria spp. The PCR technique developed is more sensitive and specific than the microbiological method of the Official Mexican Standard for the diagnosis of L. monocytogenes. Therefore, the PCR technique can be used as a complementary tool to the methodology established in the Official Standard for the detection of L. monocytogenes.











 nueva página del texto (beta)
nueva página del texto (beta)




