Introduction
Sotol (Dasylirion cedrosanum Trel) is a coalescent, perennial, and dioecious specie (there are male and female plants) (Cruz-Requena et al. 2013). In these plants, wind plays an important role during pollination. Both types of plants have a large inflorescence, where the stamens in males or pistils in females are located; the stem reaches 1 to 1.5m in height, and the leaves are slightly lanceolate, glaucous-yellow, with lateral spines. Their roots are ramified and fibrous; the head is thick and fleshy (Olhagaray et al. 2007). This plant specie grows wild in northern Mexico and in western Texas and New Mexico in the United States of America. Its economic importance lies primarily as an alcoholic beverage source, through a similar process to that used for tequila and mescal production (López 2005, De la Garza-Toledo et al. 2008, De la Garza 2010). Thus, sotol might be an important source of economic development in the short-term for the semidesert region of northern Mexico.
Dasylirion is a genus closely related to the economic, social and religious activities of semidesert Mexican communities. In these areas, sotol has had many uses as food for both humans and animals (Hellgren 1993, Urias-Silvas et al. 2008, Encina-Domínguez et al. 2013), since this plant has a high carbohydrate, protein and fiber content. The central and most tender part of the Dasylirion head was used as food by the indigenous people of Arizona USA, who cooked this structure of the plant and produced flour from dry sotol heads (Robles and España 2008). Heads from several Dasylirion species, including the central portion of the plants, and the lowest part of leaves, are good as feed for cattle during the dry season of the year, as their nutritional quality is sufficient to keep the cattle (including dairy) in good condition for long periods of drought (Powell 1998). Mancilla-Margalli and Lopez (2006) reported a chemical analysis of fresh and dry sotol heads, obtaining 26.2% and 57.2 % carbohydrates, respectively. These figures are the main reason for its use in the manufacture of liquors and spirit beverages. Cruz-Requena et al. (2013) performed a more complete analysis of Dasylirion heads and reported that they consist of 0.49 % protein, 9.63 % fiber, 5.89 % carbohydrates and 0.78% fat.
Sotol plants have been used in house and fence construction, production of alcoholic beverages (Hernández-Quintero et al. 2015), and ornaments for religious and cultural purposes. The leaves are used to make mats, hats, baskets, brooms, fire stirrers and many other objects. It has also been found that the fiber of some Dasylirion species have characteristics suitable for papermaking. The basal portion of the leaves of various Dasylirion species, which by its peculiar shape are called “spoons”, are used to decorate the interior and exterior of farm houses and villages, particularly during religious festivals. Moreover, in some northern states of Mexico, whole plants are used to decorate gardens, parks, houses, supermarkets. (Reyes-Valdés et al. 2013, Robles and España 2008). In recent times a semi-industrial exploitation as raw material for production of alcoholic beverages with the same name as the plant has begun (De la Garza et al. 2010).
Genetic diversity in the broadest sense is the most basic component of biodiversity and is defined as heritable variations that occur in each organism, among individuals of a population and the genetic structure of their populations (Wen and Hsiao 2001). Biodiversity derives from the evolutionary processes that operate on these variations. Hence, knowledge and understanding of their genetic diversity is of vital importance for conservation, advancing of evolutionary genetics, public health, sustainability, productivity of agriculture, livestock, fisheries and forestry, domestication and biomedicine (Piñero et al. 2008). The morphological, physiological and genetic variation of populations may be due to various factors such as elevation, so it is common to find different phenotypes of the same species among populations grown at different elevations (Ohsawa and Ide 2008). Therefore, the development of molecular analysis has been suggested to improve the distinction among individuals of different species or varieties (Rodríguez and Sytsma 2005, Meléndez et al. 2012). Molecular markers are powerful tools that help to study species and varieties for identification, distribution of natural diversity, and marker-assisted selection, among other uses (Robles-Esparza et al. 2012). One of the most popular markers for studying genetic diversity is the amplified fragment length polymorphisms (AFLPs). This technique is a method for detecting DNA polymorphism in a dominant way, and combining strategies of polymorphisms in the size of the restriction fragments (RFLP) and the polymerase chain reaction (Vos et al. 1995). These characteristics have made AFLPs a very useful tool in genetic diversity studies, vg, in mango (Mangifera indica L.) (Lavi et al. 1998), grape (Vitis vinífera) (Riaz et al. 2004), avocado (Persea americana Mill) (Gutiérrez-Díez et al. 2009) and cactus (Opuntia spp.) (García-Zambrano et al. 2009).
Information exists on botanical description, geographic distribution of sotol and production of the alcoholic beverage (Bogler 1998, De la Garza et al. 2010); however, information on genetics, chromosome number, DNA C value, and genetic diversity is scarce. Since Dasylirion is an allogamous plant, an elevational pollen flow between strata in a mountain is expected, and therefore high genetic diversity at the population level. This work was performed with the following objectives: to study the genetic diversity of a Dasylirion population distributed along an elevational gradient in a mountain and to determine the optimal number of individuals for sampling.
Materials and methods
Location and sampling method
Based on previous reports about geographical distribution of Dasylirion species and information provided by the National Institute of Forest, Agricultural and Livestock Research (INIFAP), Saltillo Experimental Station (Cano and Martinez 2007), the rural community “El Cinco”, Parras de la Fuente, Coahuila was selected as the sampling location, which has one of the largest sotol populations in Coahuila. This community is located at 25.1112 NL, 101.3817 WL, and 2040 masl, it has an average temperature of 14-16 °C, a hailstorm frequency of 1-2 events per year, average annual rainfall of 500-600 mm and a frequency of 40-60 d with frosts per year. Because Dasylirion plants usually grow in áreas with small mountains, slopes or foothills (Villarreal-Quintanilla and Encina-Domínguez 2005), it was decided to perform a stratied sampling, for which a sotol population growing in a mountain was divided into five strata. This population was selected because there was no evidence of plant extraction so it can be considered as an unperturbed population, and given the reproductive pattern of this species Hardy Weinberg equilibrium was assumed in the population.
Stratum one was the lowest part and stratum five was the mountain top (2040 and 2140 masl, respectively). Each stratum had a difference in elevation of 20 meters above each other. Twenty plants were sampled from each stratum. From each plant, two leaves close to the sotol head were cut; they were selected taking into account their color (gray-green characteristic of the specie), health, size (the largest leaves of the plant) and maturity (well-developed leaves without yellowing because of aging). Collected leaves were identied and stored in plastic bags.
DNA isolation
Thorns were removed from leaves; then, leaves were cut into small pieces, wrapped in aluminum foil and stored at -40 °C. DNA isolation was performed following the method described by Dellaporta et al. (1983). Tissue was weighed (0.25 g) and frozen in liquid nitrogen for 10 min; samples were transferred to a mortar previously-frozen samples were transferred to a mortar and ground to a fine powder, which was placed in a 2 mL Eppendorf tube, after which 1 mL of buffer extraction (pH 8.0 Tris-HCl 1M, 0.5M EDTA pH 8.0, 5 M NaCl, β-mercaptoethanol and 1% SDS) was added. Then, samples were placed into a water bath at 65 °C for 12 min, after which 330 µL of 5 M potassium acetate were added by gently stirring using inversions; then, samples were kept at -20 °C for 40 min. Samples were centrifuged at 16,100 g for 20 min at 4 °C, the supernatant was decanted and placed at -20 °C for 1 h in tubes containing isopropanol, and samples were centrifuged at 13,400 g at 11 °C for 10 min. At the end, isopropanol was decanted, and 200 µL of dilution solution (Tris -HCl pH 8.0 1 M, EDTA pH 8.0 0.5 M) were added to each sample along with 5 µL of RNase; samples were incubated at 37 °C for 40 min. Then, 400 µL of phenol-chloroformisoamyl alcohol (1:24:1) were added and samples were vortexed for 1 min and centrifuged for 5 min at 13,400 g; this step was performed twice. The aqueous phase was transferred into a new tube, and then 75 µL of 3 M sodium acetate and 500 µL of isopropanol were added to the sample, which later was agitated by gentle inversions, and incubated at -20 °C for one hour, then centrifuged at 9 300 g for 10 min at 4 °C; after that, the supernatant was decanted. Subsequently the pellets were washed twice with 70 % EtOH (v /v), which later was decanted, and the samples were placed vertically until etanol evaporated. Finally, DNA was re-suspended in 60 µL of 0.1 X TE. The isolated DNA integrity was assessed by electrophoresis on 1% agarose gels.
Amplified Fragment Length Polymorphism (AFLP)
First, a 1:100 dilution of each extracted DNA was performed. AFLP detection was carried out using AFLP ® Flourescent IRDye ® kit for Large Plant Genome Analysis of LI-COR Biosciences kit. Before analyzing AFLPs, different tests were conducted making combinations of 15 primer pairs to find those that offered the highest polymorphism. The AFLP analysis consisted of four stages. In the first stage, 3 µL of DNA were digested with two restriction enzymes, one with infrequent cutting (Eco RI) and the other one with frequent cutting (Mse I), and then samples were incubated at 37 °C for 2 h and at 70 °C for 15 min. After that, DNA adapters were attached to the sticky ends left by restriction enzymes using ligase. Subsequently, samples were incubated at 20 °C for 2 h. Next, a 1:10 dilution of each mixture was held for the second step (pre-amplification). At this stage, DNA was amplified through polymerase chain reaction (PCR) in a thermocycler (Thermal cycler px2, serial number: PX210642, Thermo Electron Corporation, USA) using primers with sequences complementary to the DNA adapters. At the end of the PCR, the simple was diluted to a 1:40 concentration using deionized distilled water. In the third stage, a selective PCR amplification was performed using 2 mL of the previous dilution and one primer with the termination CTG for Mse I and another primer with the termination ACA for Eco R1: both primers were labeled with IRDye 700 Fluorescence. In addition, another PCR was performed using one primer with the termination CTG for Mse I, and another primer with AGG termination for EcoR1; both primers were labeled with IRDye 800. After that, 0.5 µL of “stop” solution were added and samples were well mixed and denatured at 94 °C for 3 min. In the fourth step, amplified products were separated in a polyacrylamide gel using the IR2 4200 DNA sequencer (Li-Cor Biosciences, Lincoln, NE, USA) and finally the obtained bands were observed in the gel.
Data Analysis
The AFLP bands obtained from the polyacrylamide gels were organized in a binary matrix (0-1), where (0) represented absence and (1) presence of the band. The Info-Gen software (Balzarini and Di Rienzo, 2004) was used to determine: genetic diversity, Nei unbiased heterozygosity, average and effective number of alleles (Ne), polymorphic loci and optimal sample size to determine genetic diversity in sotol. In addition, an analysis of molecular variance was performed. A dendrogram was constructed using Info-Gen software.
Diversity was estimated using two parameters: Genetic diversity and Nei unbiased heterozygosity. Genetic diversity is a function of heterozygosity and was calculated from the sum of squares of allelic frequencies. Thus, form loci, the average genetic diversity is
Where: M= number of locus, pij is the frequency of the i and j alleles
Nei unbiased heterozygosity was estimated as follows:
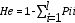 where
where 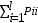 is the frequency of homozygous genotypes.
is the frequency of homozygous genotypes.
The stability of each node in the dendrogram was determined using the bootstrap test with 1000 re-samplings and Shannon entropy was calculated.
Results
Analysis of molecular variance
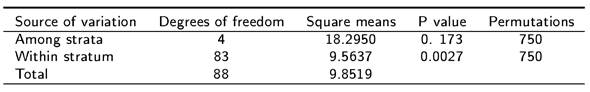
From a preliminary evaluation of six sotol plants, the combination of EcoRI-AGG and MseI CTG primers was selected based on the number and resolution of the amplified bands. This primer combination produced a total of 46 polymorphic loci in polyacrylamide gels. The amplified polymorphic bands had sizes between 50 and 700 bp. The analysis of molecular variance (Table 1) shows the existence of significant genetic variation among the five strata (P ≤ 0.05) and within each stratum (p leq 0.01).
Genetic diversity
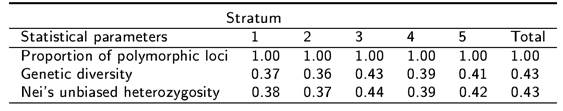
The data for each of the five strata were averaged over loci and with a confidence interval of 95 % (Table 2). AFLP data in this experiment shows that Stratum 3 had the highest genetic diversity; these results suggest that Stratum 3 (middle part of the mountain slope) had the highest genetic variation, while Stratum 1 (base of the mountain slope) had the lowest. The values determined for Shannon entropy in different strata of the sotol population followed a similar tendency to that found for the Ne (Table 2). In other words, Stratum 3 showed the highest value.
Table 2. Genetic diversity (GD), effective number of alleles (Ne), and Shannon entropy (E) for each of the elevational strata of the sotol population (p ≤ 0.05).

* =Probability. + = bits
Some genetic variability statistics calculated across all loci for each stratum are presented in Table 3. Stratum 3 is the one with the greatest genetic variation showing the highest values in heterozygosity and genetic diversity values. The probability that heterozygosity is only by chance is 0.0173 according to AMOVA (Table 1).
Principal coordinate analysis and sample size
The dendrogram obtained from the AFLP data (Figure 1) shows that there is not a group with plants from a single stratum. This is attributed to the high genetic flow among strata, suggesting that this population of plants has evolved in a similar way sharing alleles by the open-pollinated nature of this species. Figure 2 shows that the mínimum sample size to determine the genetic variability in sotol is 24-26 plants, since if sample size is increased it roughly results in the same values of proportion of polymorphic loci (a), Nei's unbiased heterozygosity (b), number of alleles per locus (c), effective number of alleles (d), and genetic diversity (e). Exceeding the optimum sample size results in a small increase in genetic diversity values and a significant increase in research costs.

Figure 1. Dendrogram of individual plants in the five elevational strata. The first number represents the stratum and the following one corresponds to the individual plant. Plants from the first stratum were numbered from 1 to 20; plants from the second stratum were numbered from 21 to 40 and so on.

Figure 2. Minimum sample size (number of plants) to calculate parameters of genetic diversity in sotol populations. a= minimum sample size to calculate polymorphic loci proportion, b= minimum sample size to calculate Nei´s unbiased heterozygosity, c= minimum sample size to calculate mean of number of alleles per locus, d= minimum simple size to calculate mean of effective alleles number, e= minimum sample size to calculate genetic diversity
Discussion
Analysis of molecular variance
The analysis of the overall AFLP amplified bands showed a 95 % of polymorphism. The power of the AFLP technique is based on the genetic variation between and within species, varieties or cultivars and it is highly sensitive for the study of genomic DNA fingerprints in any organism (Rosales-Serna et al. 2005, Meléndez et al. 2012). In this study, the variation among strata is twice larger tan the variation within stratum. This suggests that both the genetic variation generated by the horizontal pollination and that generated by the vertical pollination of plants in the mountain slope are important. Because of wind pollination and the dioecious condition of the sotol species, it is suggested that female plants are mostly pollinated by male plants from different stratum rather than by male plants from the same stratum.
Genetic diversity
Differences in polymorphism among accessions of the same species but from different locations can be attributed to the difference in allele frequencies, in particular, or to the presence of uniqueeles in genotypes in any locality (Chen and Nelson 2005). The effective number of alleles (Ae) is calculated as the reciprocal of the sum of squared allele frequencies; in other words, the classical population genetics concept of effective number of alleles at a locus, termed Ae, converts the unequal allele frequencies at a locus into a value that is equivalent to some number of equally frequent alleles (Kidd and Speed 2015). So, the effective number of alleles that can be present in a population (Balzarini et al. 2010). The Ne was larger for Stratum 3, while for other strata the values were very similar (Table 1). One possible explanation for this result is the openpollinated nature of this species, associated with the sampling site which was a hillside where monsoontype winds occur (wind direction presents a seasonal change); this factor could result in plants in the middle part of the hillside having a greater chance of receiving pollen from both the low and highlands, which is reflected in a higher proportion of alleles. It has also been reported that environmental factors such as soil, moisture stress and micro-climatic conditions influence genetic variation (Hegde et al. 2002).
Shannon parameter shows values between 0 and 1 (Greig-Smith 1983) and was used to determine how much diversity exists among strata of the sotol population according to loci diversity. When the index is zero, there is no diversity among plants in the tested stratum; in other words, the AFLP bands are all monomorphic in all plants, but when the value is larger than zero it indicates the existence of diversity among the plants analyzed according to loci diversity. Shannon entropy values were close to 1, which shows the great genetic diversity among plants of the same stratum, generated by open pollination and relevant mutations that increase the adaptive value (fitness) and create intra-population diversity (Dreisigacker et al. 2005). Information about genetic diversity allows germplasm classication within groups with contrasting variability, which is especially important in designing future breeding programs, which can be set up to develop crosses between genetically distant parents to express greater hybrid vigor than that expected from crosses between related parents (Menz et al. 2004, Meléndez et al. 2012).
Tables 2 and 3 show that Stratum 5 was the one with the second highest genetic variability; this is the stratum on top of the mountain, where soils are shallower, in this area has more lixiviation, and there is less soil moisture, thus, environmental conditions are more critical. These conditions might promote more mutations and the creation of rare alleles, resulting in an adaptation to fine scale to the ecological diversity of the region (Hegde et al. 2002). By determining the intra-specific diversity, information is obtained on behavior of this plant in its natural environment in evolutionary aspects related to its survival in adverse climatic conditions such as the semi-desert, where sotol populations normally grow (Cano and Martinez 2007).
Principal coordinate analysis and sample size
The number of plants (sample size) used to assess the genetic diversity of a population is very important, since variance of genetic diversity estimates increases with reduced numbers of sampled genotypes (Dreisigacker et al. 2005). Another factor affecting the estimation of genetic variation is population size; a positive correlation has been reported between inter-population genetic variation and population size, particularly in rare or endangered species, because small populations tend to inbreeding due to environmental effects such as fire, pruning. that can reduce their size, leading to phenomena such as founder effect or bottleneck (Martínez-Castillo et al. 2006). When sampled populations have a different size, one strategy is repeated random sampling and when similar individuals tend to cluster according to their genetic diversity, geography and/or ecotype, stratified sampling is suggested to collect samples (Wang et al. 2006).
In general, the information generated in this study may lead to better use and conservation management of sotol populations; however, it is necessary to gather more scientific information on this species, which will help to promote the greatest benefit for the inhabitants of the semi-desert region of Mexico and achieve the conservation of this specie. This generated information will allow breeders to design breeding strategies that allow them to select sotol populations with greater productive potential.
Conclusions
Based on the estimated parameters, the middle elevational level had the highest genetic variation. The optimum number of samples to determine genetic variability in sotol was from 24 to 26 plants. In the produced dendrogram, no association was observed among those plants belonging to the same stratum, which could be explained by the high genetic flow among plants from different strata.











 nueva página del texto (beta)
nueva página del texto (beta)