The most recent estimate for vascular plant and bryophyte species on Earth is ca. 403,000 with about 10 % yet to be described (Lughadha et al. 2016, Borsch et al. 2020). According to Ulloa Ulloa et al. (2017), in the Americas, there are 124,993 species of vascular plants, 6,227 genera, and 355 families, and Mexico is the country with the third highest species richness after Brazil and Colombia. Mexico has 23,314 native vascular plant species (in 297 families and 2,854 genera), of which 50 % are endemic. Mexico is the fourth most floristically diverse country in the world (Villaseñor 2016) and contributes 7.5 % of total vascular plant diversity. It has been suggested that the richness of the Mexican flora is in great part the result of unusual environmental diversity and complex orography (Rzedowski 1991). It is also influenced by the presence of three main phytogeographic elements: the Neotropical (a southern influence), the Holarctic (boreal), and the autochthonous (endemic) (Rzedowski 1991, Villaseñor et al. 2021).
There are several research strategies to arrive at a better understanding of the diversity of a region, and two basic points to address in this regard are plant identity (through checklists, floras, and monographs), and phylogenetic studies. A plant checklist represents a “verifiable list of species based on the analysis of herbarium sheets, collected specimens, published literature, and expert knowledge of plant specialists” (Ulloa Ulloa et al. 2017). A good example of a well-known checklist is The World Flora Online (WFO) project, which is a free online source of rigorously compiled and scientifically verified biodiversity data on bryophytes, ferns, gymnosperms, and angiosperms (Borsch et al. 2020). In Mexico, there are 14 ongoing floristic projects (Sosa & Dávila 1994) and one completed flora (Calderón de Rzedowski & Rzedowski 2005). Most of these projects are at the state level (local), but there are also regional projects, and there are 13 states that lack floristic inventories (Villaseñor 2016). Unfortunately, at the national level there is no flora project, though there is currently a proposal to produce the electronic Flora of Mexico, eFloraMEX (V. Sosa pers. com.).
A monograph is the systematic treatment of a plant group, which traditionally covers all the taxa corresponding to a particular category (order, family, genus), though there are also regional monographs that cover a particular region (Grace et al. 2021), and with the Tree of Life (Soltis et al. 2004) it is now possible to think about clade monographs. However, the lack of phylogenetic frameworks has limited our understanding of plant diversification and of the relationships among close species (Grace et al. 2021). Fortunately, there has been an increase in the number of phylogenetic studies that use sequencing data from open repository platforms such as GenBank (Folk & Siniscalchi 2021). Digitizing plant specimens (Soltis et al. 2018) provides, among other things, information about the geographical location of a plant species. These specimens are the principal source for compiling floras, checklists and monographs. Additionally, geographical data together with species phylogenies have led to spatial phylogenetic studies. These studies are done at the continental level (Thornhill et al. 2016, Mishler et al. 2020), the national level (Heenan et al. 2017, Scherson et al. 2017, Lu et al. 2018, Sosa et al. 2018) and the subnational level (Thornhill et al. 2017), and have used the sequencing data from GenBank to build their phylogenies.
GenBank, one of the most widely used molecular databases, is a public database of nucleotide sequences located at the National Center for Biotechnology Information (NCBI), in Bethesda, Maryland United States of America. The DNA accessions found in repositories such as GenBank commonly have biological annotations and bibliographic information in their metadata that are freely available (Benson et al. 2006). Approximately five exabytes of data were generated from the origin of civilization to 2003, and the same amount of information is currently being produced every two days (McCulloch 2013, Gupta et al. 2018). Big data are characterized by the increasing volume, variety, and velocity of data (Ford et al. 2016). Big data mining can inspire questions and generate hypotheses and hypothesis-driven science (Marx 2013, McCulloch 2013, Ford et al. 2016). Software has been developed to mine the massive amount of information stored in GenBank, including PhyLoTA Browser (Sanderson et al. 2008), phylotaR (Bennett et al. 2018b), Restez (Bennett et al. 2018a), and Datataxa (Maya-Lastra 2019). Datataxa has been successfully used to extract metadata for 2,558 species from the Flora del Bajío y de Regiones Adyacentes (Ruiz-Sanchez et al. 2019). Phylogenetic, barcoding, biogeographical, phylogenomic and diversity studies were the classifications used to mine GenBank metadata information for those species (Ruiz-Sanchez et al. 2019).
Our main goal was to search the metadata of GenBank accessions to determine the representation status of the documented vascular flora of Mexico, taking into consideration the phylogenetic and evolutionary studies that have been done at the national and international levels. By knowing the number of species that have been included in phylogenetic studies, we can identify target groups that are under-represented in this discipline and document new information that can be used for conservation policymakers to justify their proposals. By studying the number of published articles that included the Mexican flora (as well as endemic taxa), we can highlight the importance of Mexico as a biorepository. Finally, exploring the patterns in phylogenetic publications can reveal how methodological aspects have varied over time, and allows us to analyze the metrics, such as impact factor and frequency of open access, commonly used to evaluate research quality. This approach can also motivate others to plan future studies strategically, and even reconsider current policies.
For this study, we posed the following questions: (1) How many published papers have used native Mexican species in a phylogenetic context?; (2) What is the number of authors who published those papers and in which journals?; (3) What is the average impact factor of the articles published in open access journals?; (4) What are the most studied taxonomic groups?; (5) How many of all the species endemic to Mexico have sequence records in GenBank?; (6) How many phylogenetic papers include a description of new species and genera?; (7) Which phylogenetic methods and molecular markers have been used the most?; and (8) Which sequencing approaches and platforms have had the greatest impact? To answer these questions, we used the Mexican vascular plant checklist (Villaseñor 2016) as the main list of species to mine the metadata information in GenBank.
Materials and methods
Metadata extraction using Datataxa. As a baseline, we used the species reported in the Mexican vascular plant checklist (Villaseñor 2016) to identify the phylogenetic studies related to them. Then, in May 2021, we extracted all the studies from GenBank that included those species as the subjects of their analyses. To do so, we used Datataxa (Maya-Lastra 2019) and 23,314 species as input. Datataxa mined metadata information stored in GenBank accessions by doing an exhaustive search for all sequences associated with each species in the Nucleotide and the Sequence Read Archive (SRA) (NCBI Resource Coordinators 2016) databases, returning all the associated information organized in a new database. This database included the paper title, journal, author(s), publication date, and the institutions involved. The new database was further cleaned and filtered using a series of Python scripts written for this study. All documentation, scripts, and raw databases can be found in the open access GitHub repository (https://github.com/camayal/sysmexrev).
Cleaning and filtering. Commonly, the paper titles provided by GenBank contributors for each accession are not consistent. Sometimes, there are multiple titles for a single article, often with small changes in spelling. These minor changes cannot be detected by a simple duplicate search algorithm that uses exact matches. Thus, to remove duplicate titles properly, we used the Python module fuzzywuzzy (https://github.com/seatgeek/thefuzz) to identify similar titles (> 95 % of similarity) in our main database, and by using the Levenstein distances, we removed the duplicates. Then, we filtered topics using a list of terms (Appendix 1) to center our analysis on papers focused on systematics, taxonomy, and plant evolution, and kept only the titles that contained at least one of the topics. We used common words and regular expressions (regex) to look for variations in the same word, which allowed for fine cleaning. These expressions allowed us to search in two different languages, English and Spanish, as well as plurals.
Abstract manual extraction. We manually searched for all filtered titles in Google Scholar and extracted the BibTeX for every element found. To this end, we used the browser extension BibItNow! (https://github.com/Langenscheiss/bibitnow), which automates this extraction in most of the journal repositories. Repositories or files that did not allow automatic BibTeX creation were processed manually. In BibTeX format, we included the corrected paper title and authors, year, journal information, and abstract. Manual adjustments were needed for some dates and authors’ names with non-ASCII characters.
Thematic extraction and descriptive statistics. We used the bibtexparser module for Python (https://github.com/sciunto-org/python-bibtexparser) to manage all BibTeX citations and to conduct our thematic search in abstracts, titles, journals, and authors. This search was divided into two categories: simple regular expression searches and more advanced searches. In simple searches, we looked for the occurrence of one or more terms associated with a particular topic as described in Appendix 2. For topics involving external databases, we wrote scripts to collect that information and perform advanced searches. We used the databases from SCImago Journal Rank (www.scimagojr.com) from 1999 to 2020 to obtain information about the journal in which each paper was published, such as open source, indexation in SciELO’s list, publication country, quartile, and impact factor.
To report on endemic Mexican species studied in the papers gathered for the present analysis, we used an independent database that includes all of the species mentioned by Villaseñor (2016). Due to subtle differences in classification systems between Villaseñor (2016) and GenBank, we unified all our taxonomic reports based on the former; this applies to the families and divisions included in this paper.
To determine the frequency of use of genetic markers through time, we built a Python dictionary with regular expressions (Appendix 2) to search for the most commonly used markers over the last three decades (Clark et al. 1995, Soltis et al. 1997, 1998, Källersjö et al. 1998, Fishbein et al. 2001, Shaw et al. 2005, 2007, Smith & Brown 2018). Similarly, we extended this search to evaluate the frequency of use for all three sequencing platforms over time. We defined the three generations as follows: First generation sequencing includes all of the studies that used genomic data sequenced with the Sanger method such as AFLPs, Microsatellites, and nuclear, chloroplast and mitochondrial markers. Second generation sequencing included a broad spectrum of technologies such as 454, Illumina short-read, and Ion torrent sequencing, as well as some library preparation techniques such as RAD-Seq, Hyb-Seq (i.e., Angiosperm353 probe set). Finally, for third generation sequencing we included only PacBio and Oxford Nanopore technology (Slatko et al. 2018).
The final database is available at the following public repository https://doi.org/10.5281/zenodo.5651811
Results
Native Mexican species used in phylogenetic studies. Datataxa extracted metadata associated with 12,589 species in the Nucleotide database and 2,026 species in the SRA database. From both datasets, 12,168 and 4,035 unique titles were gathered, respectively. In the SRA database, most of the titles were only the name of the species, variety, or cultivar, whereas in the Nucleotide database, most of them represented the actual title of a published paper. Only 4,991 (41.02 %) and 153 (3.79 %) titles were maintained after filtering by topic (systematics, taxonomy, and plant evolution). After the manual extraction of the abstracts, the total number of papers in the final database was 3,807. Records for these papers contain the title, abstract, authors, journals, and other bibliographic information such as DOI, ISSN, publication date, volume, and pages. The remaining 1,337 titles were excluded because they were not articles, book chapters, or other citable written works.
Journals and authors. Scientists published phylogenetic results on Mexican plants in 325 different peer-reviewed journals. In the search performed, there are at least three recognizable journal groups. The first group includes three journals with more than 400 articles, with Systematic Botany standing out at 457 published papers. The next group includes journals with more than 100 articles and fewer than 300 articles, in which Taxon has 270 published papers (Figure 1A). Finally, are all the journals with fewer than 100 articles. In this category, it is worth mentioning that about 299 journals have fewer than ten published papers. In this last group are the national journals (Botanical Sciences and Acta Botanica Mexicana) with five between the two on this topic. The number of authors of published papers varies most frequently between 1 and 6 (Figure 1B).

Figure 1 Top journals with the most manuscript published that have used native Mexican species in a phylogenetic context and the dispersion of the impact factor. (A) Bar chart showing the number of manuscripts published in the first 30 journals. (B) Heatmap plot showing the number of manuscripts published considering impact factor and number of authors. (C) Box plots showing the number of manuscripts published considering impact factor and year of publication. The dotted line represents the mean for the entire dataset.
Impact factor and open access journals. Of 3,807 papers, 93 % were published in a journal listed in the SCImago database. Of those, only 374 (11 %) were open access. Of the total set of papers published in journals indexed in SCImago, just 14 papers (0.36 %) were published in SciELO (bibliographic database: the Americas, Iberian Peninsula, South Africa), all of which were open access. The mean impact factor was 1.64 (SD = 1.33; median = 1.42; max = 19.69; Figure 1C).
Taxonomy, endemic species, and new taxa. At the division level, the largest number of publications were on Magnoliophyta (+3,300), followed by Polypodiophyta and Coniferophyta (Figure 2A). At the order level, each of the 70 orders reported in Mexico has at least one published paper. Poales and Lamiales were represented by more than 300 articles each, followed by Fabales, Asterales, and Asparagales with more than 200 papers each. Together with five other orders, they account for more than half of the publications (Figure 2B). At the family level, the greatest species diversity is reported for Poaceae, Fabaceae, Asteraceae, Solanaceae and Orchidaceae, with more than 100 publications each (Figure 2C). Finally, we found 17 families (Achariaceae, Balsaminaceae, Cunoniaceae, Dichapetalaceae, Mayacaceae, Mitrastemonaceae, Monimiaceae, Musaceae, Nitrariaceae, Opiliaceae, Peraceae, Resedaceae, Schoepfiaceae, Tapisciaceae, Theaceae, Vochysiaceae, and Woodsiaceae) that, despite having at least one paper in our database, did not include any Mexican species in the ingroup. Of the 23,314 species that occur in Mexico, 43 % (10,094 species) have at least one report in GenBank. Considering only those that are endemic to Mexico (12,013 species), a total of 3,301 species (27 %) have been used in 1,363 papers analyzed in this study. Additionally, of the more than three thousand articles obtained in our search, 123 focus on the description of new species and 83 on new genera (Figure 3A).
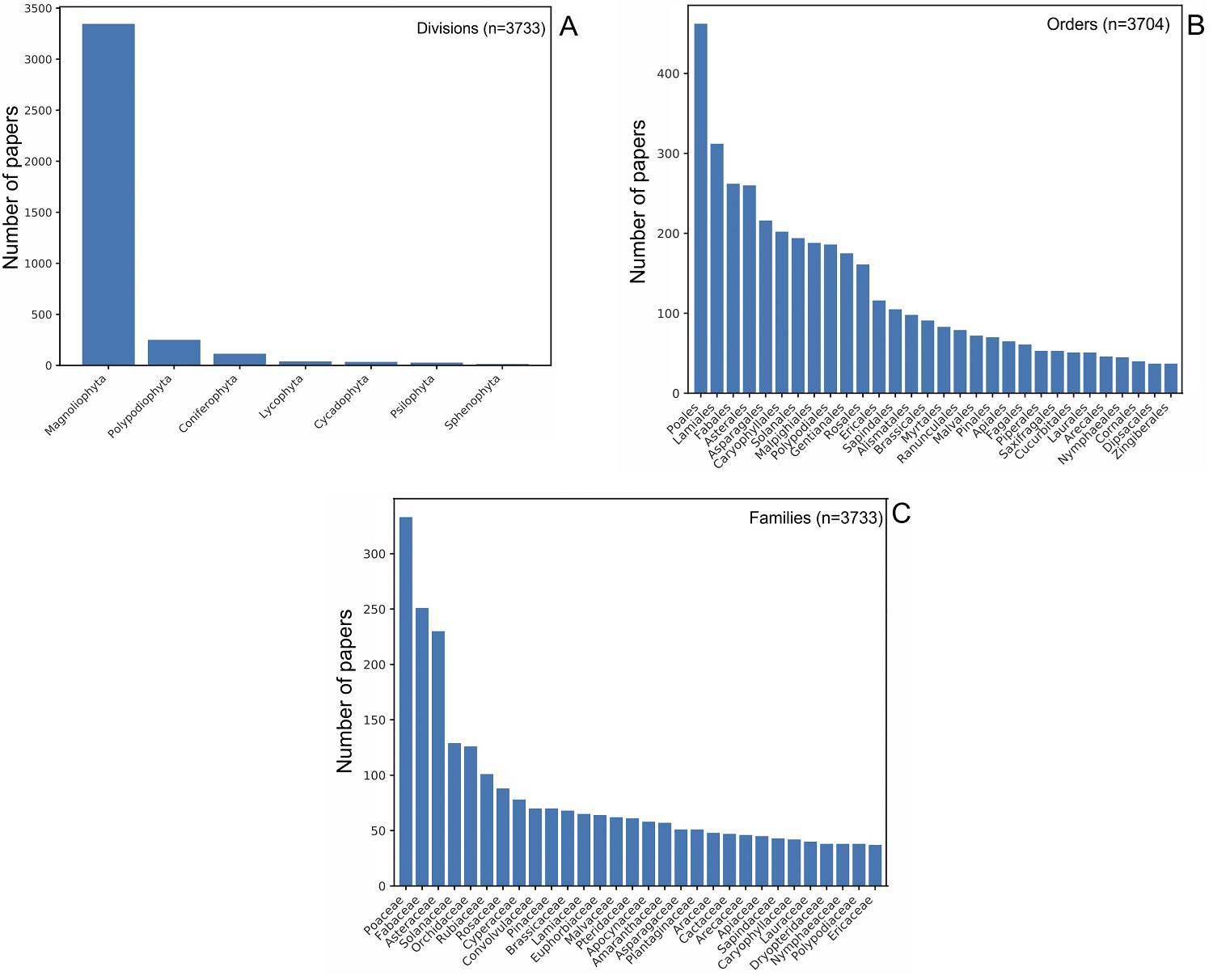
Figure 2 Trend in manuscript publication according to different taxonomic categories. (A) Bar chart showing the number of manuscripts published taking into account the division taxonomic category. (B) Bar chart showing the number of manuscripts published considering the order taxonomic category. (C) Bar chart showing the number of manuscripts published taking into account the family taxonomic category.

Figure 3 Trend in the publication of new species and phylogenetic methods in the last 20 years. (A) Bar chart showing the number of manuscripts published that include descriptions of new genera and new species per year. (B) Bar chart showing the number of manuscripts published considering Parsimony, Maximum Likelihood, and Bayesian Inference phylogenetic methods per year.
Phylogenetic methods. Since 1990, the phylogenetic inference methods used to analyze molecular data for the Mexican flora are Maximum Parsimony (MP), Maximum Likelihood, and Bayesian. The MP method is historically the most employed but is currently the least used. The second is Bayesian inference, whose use began in 2000 and is now the most widely applied, and in third is ML (Figure 3B).
Molecular markers. Regarding the genomic source of the molecular markers used -chloroplast vs. nuclear- 2,320 publications on the Mexican flora used chloroplast markers, and 2,096 publications used nuclear markers (Figure 4A). We identified at least 30 markers (Figure 4B). Of these, only nine markers are found in more than 100 publications. We can classify these markers into groups based on their degree of use. The first group is composed of ITS (ribosomal), which is the most used marker and was reported in more than 1,200 papers. The second group of markers is composed of three chloroplast markers (trnL-trnF, rbcL, matK) with more than 500 publications. In the third group of markers, there are plastid (ndhF, trnH-psbA), nuclear (ETS), and ribosomal (18S rDNA) markers that have been used in more than 100 studies. The fourth group includes the remaining 21 markers (Figure 4B).

Figure 4 Trend in the use of molecular markers (nuclear and chloroplast) and sequencing platforms in the last 20 years. (A) Bar chart showing the number of manuscripts considering the source of the markers (chloroplast and nuclear) per year. (B) Bar chart showing the number of manuscripts published taking into account the sequencing platform (first = Sanger sequencing, second = RADseq, Angiosperm353, hybseq, 454, and third generation = nanopore, PacBio) per year. (C) Bar chart showing the number of manuscripts considering the top 30 most used molecular makers.
Sequencing platforms. We found three types of sequencing directly associated with markers. ‘First generation sequencing’ (i.e., Sanger sequencing) was the most used and reported in more than 2,700 studies, followed by ‘2nd generation sequencing’ (i.e., 454, Illumina short-read, and Ion torrent sequencing) with 169 articles and a peak of 40 articles in 2020, and ‘3rd generation sequencing’ (i.e., PacBio and Oxford Nanopore) with just three reports (Figure 4C).
Discussion
Representation of native Mexican species in a phylogenetic context. The quest for knowledge of the national flora is one that is still under development. With the application of phylogenetic inference tools and the use of molecular markers and, in some cases, morphological data, the study of this flora has brought significant advances in both the discovery of taxa and also the relationships between different taxa. Our results show that from about 4,000 articles, little more than 40 % of the floristic species diversity has been covered, though there is an upward trend in the proposal of new species and genera using molecular markers and phylogenetic inference tools (Figure 3A). With 122 articles per year involving Mexican species, we can suggest that there has been a notable advance in phylogenetic analysis over the last 30 years. The scope remains to be seen, but we now know that including terminals in the phylogeny will allow for a better understanding of the processes and the evolution of the groups at different geographical and taxonomic scales (Blackburn et al. 2019, Rivera et al. 2021).
Sosa et al. (2018) published the first study on the spatial phylogenetics of the Mexican vascular flora and the phylogenetic tree consisted of 9,731 terminals (species), just over 40 % of the Mexican flora. This tree was based on the calibrated phylogeny of Zanne et al. (2014), which included seven (18S rDNA, 26S rDNA, ITS, matK, rbcL, atpB, and trnL-F) gene regions downloaded from GenBank. Our results showed that ITS, matk, rbcL and trnL-F are among the most used markers in phylogenetic studies. To complete the phylogenetic tree of the Mexican vascular flora, we must sequence more Mexican endemic species with the same molecular markers that Zanne et al. (2014) used in their phylogenetic analysis.
Higher taxonomic level taxa in GenBank. Our results at the division level showed that Magnoliophyta has at least 13 times more published papers than any of the other divisions (Figure 2A). This tendency is most likely correlated with the fact that the highest diversity in Magnoliophyta occurs in Mexico (Villaseñor 2016). At the order level, we found that Poales, Lamiales, Fabales, Asterales, and Asparagales are the first five orders with the most published papers with records in GenBank (Figure 2B). We found the same orders as Villaseñor (2016), but Poales was first in our results. Several species of grasses, such as Zea mays L., have been used as a model for genetic studies. The studies that have used several species of grasses or the great number of species that this order has may explain why Poales is so well represented in GenBank.
Finally, at the family level, we found that Poaceae, Fabaceae, Asteraceae, Solanaceae, and Orchidaceae have the most published papers with records in GenBank (Figure 2C). According to Ulloa-Ulloa et al. (2017), in the Americas, Orchidaceae, Asteraceae, and Fabaceae are the three most species-rich families. Meanwhile, at the national level (Mexico), Asteraceae, Fabaceae, Orchidaceae, Poaceae, and Euphorbiaceae are the top five families with the highest diversity (Villaseñor 2016). At the order level, the grasses used as model species could increase the records in GenBank. In our results, we found that Solanaceae was the family with the fourth highest number of records. This group has been extensively studied, and in our analysis we found 129 studies that included 227 species. This could be due to the economic importance of potato (Solanum tuberosum L.) and tomato (Solanum lycopersicum L.) species as plant models in several genetic studies.
The most diverse families have consolidated national or international working groups (e.g., compositae.org; Grass Phylogeny Working Group 2001) that contribute a substantial quantity of data to GenBank. Of the 297 families included in Villaseñor’s list, 30 account for almost 70 % of the phylogenetic articles, after which the number of contributions for other groups decreases rapidly. Is there a bias because there are more people working in diverse groups? Does this reflect the taxonomy crisis in the country? Perhaps a bit of both, since many taxonomists are working in groups devoted to high diversity taxa (Directory of Taxonomists 2019, Botanical Society of Mexico, www.socbot.mx/documentos.html); even so, there are not even enough taxonomists to cover those groups. For example, we detected at least 11 people working with Orchidaceae, 12 with Fabaceae, nine each with Poaceae and Cactaceae, and seven with Asteraceae. However, over this period, the incorporation of new taxonomists has not been as fast (Villaseñor 2015), nor are they focusing on groups with lower levels of diversity.
Endemic Mexican species in GenBank.Villaseñor (2016) recorded 23,312 vascular plant species that occur in Mexico, of which 12,013 are endemic. Two years later, Sosa et al. (2018) recorded a higher number of vascular plant species for Mexico (24,630), of which 10,235 are endemic. We found that 10,094 species have at least one record in GenBank, and of the endemic species only 3,301 have one or more records in GenBank. Among the species reported by Sosa et al. (2018), only 1,664 species were used to build their phylogeny. Regardless of which checklist is used, Villaseñor’s (2016) or Sosa et al.’s (2018), the fact is that endemic Mexican species with usable records in GenBank are significantly under-represented when building a phylogenetic tree of the Mexican vascular plant species.
New taxa described in phylogenetic papers. We recorded 123 papers that included descriptions of new taxa since 2000, which used molecular markers deposited in GenBank and include Mexican species (Figura 3A). According to Villaseñor (2015) and Alvarado-Cárdenas et al. (2021, 2022), each year around 100 new species are published for Mexico. In 2020, 105 taxa, and in 2021, 80 taxa were described from Mexico. Ninety (2020) and 76 (2021) of the newly described species are endemic to Mexico (Alvarado-Cárdenas et al. 2021, 2022). The five families with the most records in GenBank are Poaceae, Fabaceae, Asteraceae, Solanaceae, and Orchidaceae (Figure 2C). However, among the families with a greater number of described species in Mexico during 2020 were Piperaceae, Bromeliaceae, Fabaceae, Convolvulaceae, and Apocynaceae (Alvarado-Cardenas et al. 2021). The low representation of new species records in GenBank published for Mexico could result from several factors. One is the use of morphological characters in species descriptions instead of molecular data via phylogenetic species delimitation analyses. Another reason is that many species are described from herbarium specimens, collected 11 to more than 100 years ago (Bebber et al. 2010, Villaseñor 2015), from which it can be difficult to extract DNA.
For genera, 83 of them were described in the period analyzed (not limited to Mexico). These data are difficult to compare because, unlike the situation with species, there is no similar evaluation. We can say that from 1998 to 2021, an average of 3.6 papers were published per year identifying new genera. The years 2014 and 2020 stand out for having the highest number of papers, with eight and 15 new genera, respectively. Villaseñor (2004) mentioned that in 1946 Recko had reported 2,189 genera of vascular plants for the country and by 2004, this number had grown to 2,804 (a difference of 615 genera, equivalent to 9.5 new taxa per year) and for 2016, 2,854 were reported (4.1 new taxa per year for this period). These increases at genera level could mainly be due to nomenclatural changes and others had not been previously reported. We can noticed a potentially increasing trend in the publication of these taxa.
Journal, open access, and impact factor patterns. The impact factor calculated by Thomson Reuters takes the number of citations of papers published by the journal in the previous two years and divides it by the number of papers published by the journal in that period (Simons 2008). We found that most of the papers with records in GenBank were published in journals with impact factors ranging between 0.1 and 3.3, with a mean of 1.64 (Figure 1). The top five journals in which Mexican species were included are Systematic Botany, Molecular Phylogenetics and Evolution, American Journal of Botany, Taxon, and Plant Systematics and Evolution (Figure 1A, B). None of the Mexican journals appear in the top 30. This could be because Mexican journals did not obtain impact factor status until 10 years ago. Although the impact factor is a bibliometric indicator (Kumar 2018), it is increasingly being used to evaluate papers, scientists and institutions (Simons 2008), and Mexico is no exception. We found that most of the papers published have between 1 and 6 authors (Figure 1B). However, there has been an increase in the number of contributing authors over time (Wren et al. 2007), which could result from papers being used as evaluation criteria for promotion, tenure or funding, as Wren et al. (2007) hypothesized; with more collaborative papers covering a larger scope.
We found that the number of papers published in open access journals (as defined in the SCImago index) represent the same percentage found a decade ago for science in general (Laakso & Björk 2012) and half of the number currently published in open access (Björk & Korkeamäki 2020). There is a strong tendency to publish in journals that are not open access; in fact, the most frequent journals found in this meta-analysis only offer open access “on demand” rather than the default publication policy. Offering hybrid or “on demand” open access options may help solve this lack of openness; however, the cost associated with publishing in this modality is exorbitant for those in developing economies, to the point of becoming prohibitive for researchers based in those countries. For example, in Mexico the minimum daily wage (MDW) for 2021 was around 10.56 USD (213.39 MXN; CONASAMI 2021), yet the publication cost in any of the top four journals varied from 47 MDW in Systematic Botany to 340 MDW in Molecular Phylogenetics and Evolution, i.e., an entire year of Mexican minimum wages. The other two top journals are published by John Wiley & Sons, Inc., which charges around 142 MDW for some open access options. Even though free open access journals are available, they are not attractive to researchers due to their low impact factor. We found that 14 papers (0.36 % of all papers studied here) were published in free open access journals indexed by SciELO, but their mean impact factor was 0.349 (SD = 0.21). This impact factor is not high enough for a researcher to get promoted in Mexico’s Sistema Nacional de Investigadores (SNI; The Mexican National System of Researchers) (CONACYT 2021), which requires a minimum impact factor of 0.5 for most articles presented by researchers listed in Area II (Biology and Chemistry). Most papers are being published in journals with an impact factor < 1.6 (Figure 1B).
Tree inference methods, sources of information, and sequencing technologies. There are several methods for inferring phylogenetic relationships among taxa (Brocchieri 2001). The methods most used today are Maximum Likelihood and Bayesian Inference (Duchen 2021). Our results for the preferred phylogenetic methods used from 1990 to the present for studying molecular data from the Mexican flora are Maximum Parsimony with 1,000 papers, followed by Bayesian Inference with 721 papers and Maximum likelihood with 450 papers (Figure 3B). We observed a tendency toward using statistical inference methods (Bayesian Inference and Maximum Likelihood) over Parsimony through time. Additionally, probabilistic methods have implemented numerous new models that attempt to solve different problems associated with the evolution of markers. It has also been suggested that the results were the most consistent, accurate, and robust under distinct conditions (Soltis et al. 2004, Brandley et al. 2009, Vernygora et al. 2020).
The primary sources of molecular markers used to study the Mexican flora are chloroplast with 2,320 papers, followed by nuclear markers with 2,096 papers (Figure 4A). We found a tendency to increase the use of ‘second- generation sequencing’ over the last five years (Figure 4C). However, the number of papers published that used ‘first-generation sequencing’ (Sanger sequencing) is 17 times greater than those using ‘second-generation sequencing’ (2,773 papers vs. 163 papers) (Figure 4C). Among the sources of available molecular chloroplast and nucleus markers, we found that ITS had double the number of published articles relative to any single marker of the chloroplast genome (Figure 4C). Among the five most-used markers in published papers of the Mexican flora are: ITS, trnL-trnF, rbcL, matK, and ndhF (Figure 4B). Shaw et al. (2005, 2007) published a list of non-coding chloroplast markers that are useful for inferring phylogenetic relationships to the inter- or intraspecific level. Our results revealed that some of the markers evaluated by Shaw et al. (2005, 2007) have been used less than the classical markers have (ITS, rnL-trnF, rbcL, matK, and ndhF) in the study of the Mexican flora. This probably results from the fact that classical markers are used to resolve phylogenetic relationships above the genus level, and those proposed by Shaw et al. (2005, 2007) were mostly applied below the genus category or in intraspecific studies.
After reviewing 3,807 abstracts published in 325 peer-reviewed journals, we only found five articles published in two Mexican scientific journals. We encourage the editors of Mexican scientific journals such as Botanical Sciences, Acta Botanica Mexicana and Revista Mexicana de Biodiversidad to look for alternatives to promote the publication of systematics articles in their journals and increase the number of Mexican species in international repositories such as GenBank. The impact factor is a decisive element at the moment in searching for a journal, motivating authors to send their manuscripts to journals with higher indexes. However, the aforementioned journals are very close to reaching an impact factor of 1.0, exceeding the threshold established by CONACYT in the evaluation of researchers. Encouraging researchers to publish in these journals could also increase the number of publications in Spanish, the third most widely spoken language in the world.
Finally, we have explored the importance of Mexico as a biological repository for understanding the evolution of plants in global science. Although there has been robust sampling in different taxonomic categories, further sampling is necessary for many of the groups in this mega-biodiverse country, mainly for endemic species that can contribute to the resolution of different lineages in the phylogeny.











 nueva página del texto (beta)
nueva página del texto (beta)



