In the past decades our knowledge of phylogenetic relationships within members of the Bignoniaceae has improved substantially thanks to phylogenetic reconstructions based on molecular data of the entire family (Spangler & Olmstead 1999, Olmstead et al. 2009), its main tribes (Zjhra et al. 2004, Lohmann 2006, Grose & Olmstead 2007a, Li 2008, Callmander et al. 2016, Ragsac et al. 2019), or key genera (Kaehler et al. 2012, 2019, Fonseca & Lohmann 2015, Medeiros & Lohmann 2015, Fonseca & Lohmann 2018, Thode et al. 2019, Carvalho-Francisco & Lohmann 2020). These phylogenetic reconstructions formed the basis for a series of new taxonomic treatments for the family (Grose & Olmstead 2007b, Lohmann & Taylor 2014). However, the phylogenetic placement of taxa that combine a narrow distribution and a rather ambiguous morphology has remained uncertain (Pace et al. 2016). Astianthus D.Don is one of such examples. This monotypic genus only includes Astianthus viminalis (Kunth) Baill. (Figure 1), a species distributed across the Pacific Coast side of central and southern Mexico and northern Central America (El Salvador, Guatemala, Honduras, and Nicaragua), typically associated to riparian habitats (Gentry 1980, 1992). Because of its amenable stature, attractive perennial foliage, and intense flowering, A. viminalis is sometimes planted as an ornamental along streets in southern Mexico. This species is also used in medicine, especially to treat diabetes (Meckes et al. 2001, Pérez-Gutiérrez et al. 2009).
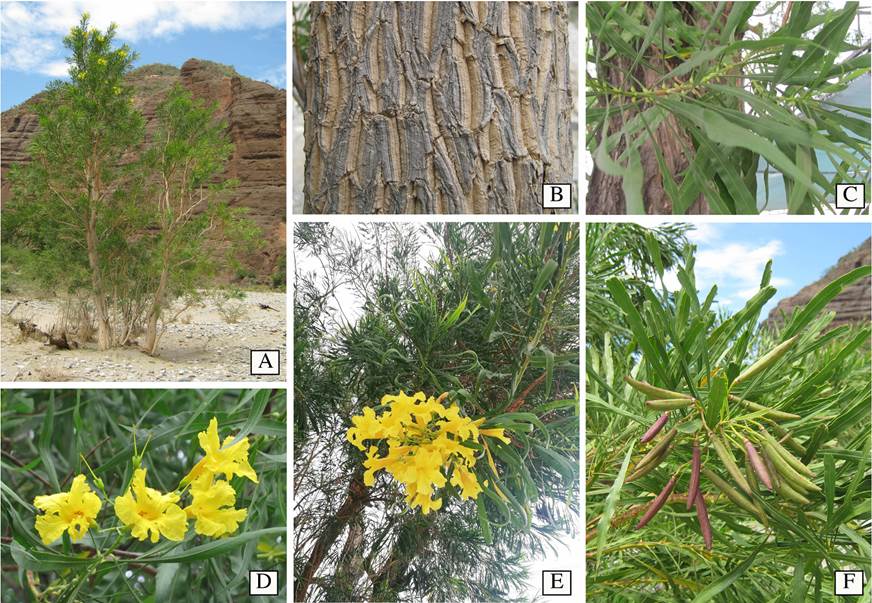
Figure 1 General morphology of Astianthus. A. Plant habit, a tree approximately 15 m high, growing in Cuicatlán, Oaxaca, Mexico. B. Rough bark. C. Simple, pulvinate, entire, verticillate leaves. D. Yellow, campanulate flowers, calyx with five acute triangular teeth, and tubular infundibuliform corolla. E. Terminal, paniculate inflorescence. F. Reddish terete capsules in a terminal infructescence. Image credits: A-B, F Esteban Martínez; C-D Carlos Cavazos; E, Carlos Domínguez-Rodríguez.
Regardless of the economic importance detailed above, Astianthus taxonomic placement has remained controversial due to its unusual morphological features such as the verticillate pulvinate, simple leaves (Figure 1C), and the 4-loculed ovary derived from the formation of a false septum in addition to the regular septum found in other Bignoniaceae (Gentry 1980, 1992). Within the Bignoniaceae, verticillate, simple leaves are found elsewhere in members of tribe Catalpeae, such as Catalpa Scop. and Chilopsis D. Don, and a few other scattered genera or individual species across the family. Two other genera that share these traits, Deplanchea Vieill. and Delostoma D.Don, were previously placed in the Tecomeae s.l., While the former is currently included in Tecomeae s.s. (Olmstead et al. 2009), the latter remains unplaced as it has emerged with low support as its own single lineage and sister to the bulk of the Bignoniaceae (Olmstead et al. 2009). Four-loculed ovaries are also rare in the family, and are only known to occur in two other genera: Tourrettia DC., a herbaceous vine in the Andean tribe Tourrettieae, and Heterophragma DC., an Asian tree, previously included in the Tecomeae s.l. (Fischer et al. 2004), but currently placed within the Paleotropical clade (Olmstead et al. 2009).
In addition to the simple, pulvinate, entire, verticillate leaves (Figure 1C, E-F) and a 4-loculed ovary, Astianthus viminalis is also recognized for being a tree with rough bark (Figure 1A-B), 10 to 25 m high, generally occurring in dense populations (Gentry 1992) - the species is sometimes also described as a shrub (Fischer et al. 2004). Since Astianthus commonly grows near rivers and streams, flowering and fruiting branches sprouting from subterranean stoloniferous roots can result in a shrubby aspect (as observed by E.M.M.S.). Astianthus has terminal, paniculate inflorescences (Figure 1E), campanulate flowers, 5-dentate calyces (Figure 1D), and tubular-infundibuliform yellow corollas (Figure 1D). The capsular fruits are reddish-green, terete, fusiform, and glabrous (Figure 1F), with winged seeds borne perpendicularly on the septum and parallel to the false septum (Gentry 1992).
The genus Astianthus was initially placed by De Candolle (1838) in the Eubignonieae, a group that contained all Bignoniaceae with septicidal capsules (i.e., fruit dehiscence parallel to the septum). This placement was likely due to Astianthus’ false additional septum and the genus was subsequently transferred to Catalpeae sensuDe Candolle (1845). This tribe included the Bignoniaceae with loculicidal capsules (i.e., fruit dehiscence perpendicular to the septum), consistent with its real septum. Also based on the loculicidal capsules, Bentham & Hooker (1876) subsequently transferred Astianthus to Tecomeae, a classification continued by Gentry (1992).
As currently circumscribed, Tecomeae s.s. includes 12 genera distributed worldwide, in both the Northern and the Southern hemispheres, while other members of Tecomeae s.l. are now placed within Catalpeae, Jacarandeae, the Paleotropical clade (including the Malagasy tribe Coleae), and the Neotropical Tabebuia alliance (including tribe Crescentieae) (Olmstead et al. 2009). Based on the Neotropical distribution and morphology (e.g., simple, verticillate leaves, and loculicidal capsule), Astianthus seems to fit best within Catalpeae, an exclusively North American tribe resurrected from De Candolle’s Prodromus (1845). However, Astianthus has a false septum, a feature not found in any Catalpeae. The morphological similarity between Astianthus and members of Catalpeae, especially the sympatric Chilopsis, was noted previously (Gentry 1992). However, Gentry (1992) noted that Astianthus was even more similar to Tecoma Juss., with which it shares similar flowers, fruits, and geographical distribution. On the other hand, Tecoma is nowadays included in Tecomeae s.s. which predominantly includes lianas, shrubs, and treelets with pinnately compound leaves (Olmstead et al. 2009), although some species are trees with simple leaves (Gentry 1992).
In addition to molecular systematic studies, wood anatomy has been central in the understanding of taxonomic affinities within Bignoniaceae and delimiting synapomorphies to different lineages (Pace et al. 2009, 2015a, b, 2016). Even in pre-molecular times, the presence of variant secondary growth was recognized as unique to tribe Bignonieae (Crüger 1850, Schenck 1893), and this feature was used to circumscribe species in and out of this tribe (Gentry 1980, Lohmann 2006). Also, wood anatomical differences previously described for large genera of Bignoniaceae were later shown to match clade subdivisions found in molecular studies. The genus Tabebuia s.l. is a prime example. It was extensively studied by wood anatomists given its economically important timber (Record & Hess 1943), and three very distinctive groups were established based on their wood characters: (i) those very hard, durable Tabebuia Gomes ex DC. woods used in carpentry, (ii) those of light wood used in the transport of fruit and vegetables locally called as caixeta, and (iii) those with woods anatomically intermediate between the previous two groups (Record & Hess 1943, Dos Santos & Miller 1992). A molecular phylogenetic study provided additional support for these same three groups (Grose & Olmstead 2007a), leading to a new generic classification (Grose & Olmstead 2007b). Under the new system, the species with light wood were maintained in Tabebuia, while those with hard wood were transferred to Handroanthus Mattos, with some species of intermediate woods included in Roseodendron Miranda (Grose & Olmstead 2007a, b). Not all species of intermediate woods have yet been studied to date in order to verify their taxonomic placement. Other examples of wood anatomical traits being of phylogenetic value within the Bignoniaceae abound: the differences in ray width and composition are useful in delimiting two main clades within tribe Jacarandeae. In this latter, it is remarkable that Jacaranda copaia (Aubl.) D.Don, the only species with anatomically intermediate wood, forms a separate lineage (Dos Santos & Miller 1997, Ragsac et al. 2019). There are also wood characters that consistently help delimit the clades most similar to Astianthus, i.e., Catalpeae and Tecomeae s.s. The Catalpeae has unique simple to semi bordered pits and abundant tyloses, while Tecomeae s.s. has the unique combination of rays with body cells procumbent and marginal cells square to upright, a tendency to a storied structure, and scanty paratracheal to aliform parenchyma (Pace et al. 2015a).
Given the importance of both molecular phylogenetic data, wood and bark anatomical characters for Bignoniaceae systematics, we combine both types of evidence to unravel the enigmatic phylogenetic placement of Astianthus.
Material and methods
We collected samples of Astianthus viminalis for anatomical and phylogenetic studies in the field. To broaden our geographic sampling, we included additional samples of A. viminalis from the National Herbarium of Mexico and its wood collection (MEXU; see Table 1). To determine the phylogenetic placement of Astianthus, we generated sequences of A. viminalis, as well as sequences of Bignonia potosina (K. Schum. & Loes.) L.G. Lohmann, and Tecoma stans (L.) Juss. ex Kunth for this study (see Table 1 for collection or specimen information). All other sequences of ingroup and outgroup taxa were the same as those used in (Olmstead et al. 2009; a complete list with voucher information and GenBank accession numbers can be found in this study). We also included 25 additional sequences from taxa available on GenBank (Supplementary material 1, Tabla S1) to fill gaps in rbcL sequences from the original sampling.
Table 1 Specimens of Astianthus viminalis and relatives which were exclusively sampled for this study. Herbarium abbreviations follow Thiers (2017). All other species sampled and their complete collection information can be found in Olmstead et al. (2009).
| Species | Collector and number |
Locality in Mexico | Used in molecular analyses |
Used in anatomical analysis |
Herbaria |
|---|---|---|---|---|---|
| Astianthus viminalis | M.R. Pace 895 | Oaxaca, Huatulco, La Crucecita, Lecho del Río |
Yes | Yes | MEXU, SPF, MO, US |
| Astianthus viminalis | J. Barajas Morales 408 | Jalisco, La Huerta, Estación Biológica de Chamela |
No | Yes | MEXU |
| Astianthus viminalis | J. Barajas Morales 531 | Puebla, Coxcatlán, Lecho del Río Calipan |
No | Yes | MEXU |
| Astianthus viminalis | J. C. Soto N. 18603 | Guerrero, Zirándaro, cauce del Río del Oro |
Yes | No | MEXU |
| Astianthus viminalis | C. Rojas-Martínez 107 | Puebla, Río Tizac, selva baja caducifolia |
Yes | No | MEXU |
| Bignonia potosina | M.R. Pace 818 | Tabasco, Balancán, Margen del Río Usumacinta |
Yes | No | MEXU, SPF |
| Tecoma stans | M.R. Pace 906 | Ciudad de México, Instituto de Biología, Ornamental |
Yes | No | MEXU |
DNA extraction, amplification, and sequencing. We extracted DNA using the DNeasy Plant Mini Kit (Qiagen, Valencia, California) from either silica-dried plant material, fresh material, or herbarium specimens, following the manufacturer’s protocols.
For each accession, we amplified portions of the ndhF and rbcL genes and the trnL-F spacer. These three regions from the plastid genome have been useful to estimate phylogenetic relationships within the Bignoniaceae at the tribal (Olmstead et al. 2009), generic (Lohmann 2006), and species (e.g.,Fonseca & Lohmann 2015, Carvalho-Francisco & Lohmann 2020) levels. We sequenced the ndhF marker in two pieces, using the PCR primer pairs 5F-1318R and 972F-3R described in an earlier study (Olmstead & Sweere 1994). For the trnL-F region, we used primers C and F (Taberlet et al. 1991), and for rbcL, we used F and R primers previously described (Hipkins et al. 1990, Supplementary material 2, Figure S1). PCR reactions were prepared by adding each primer at 1mM to GoTaq® Green Master Mix (Promega M1722), and adjusting with ddH20 for a final volume of 25µL. PCR conditions consisted of an initial denaturation of 90s at 96 ºC, followed by 35 cycles of 30s at 95 ºC, 60s at 55 ºC, 60s at 72 ºC, and a final extension of 4min at 72 ºC. Upon completion, PRC products were run in a 1% agarose gel and assessed for size and quality in a UV transilluminator using gel red (Biotium 41003, Hayward, California).
Sanger sequencing was carried out at the National Biodiversity Laboratory (LaNaBio) at the Institute of Biology, UNAM, Mexico, in both directions. Briefly, sequencing reactions were prepared using BigDye Terminator 3.1 (Applied Biosystems), and run for 30 cycles consisting of 10s at 96 °C, 5s at 50 ºC, and 4 min at 60 ºC. Samples were cleaned using Centri-Sep plates (Thermo Fisher Scientific, Waltham, Massachusetts) following the manufacturer’s directions. Samples were analyzed in an Applied Biosystems ABI 3730xl 96-capillary DNA analyzer (ThermoFisher Scientific, Waltham, Massachusetts).
Sequence editing and alignment. We examined and edited raw chromatograms for all newly generated sequences in Sequencher 5.4.6 (Gene Codes, Ann Arbor, Michigan). Contigs were assembled using default settings without lowering the threshold. We aligned each region individually and manually in Mesquite 3.5 (Maddison & Maddison 2011), giving preference to transitions over transversions. All sequences generated for this study are available in GenBank with the following accession numbers: MT235272-MT235276 (rbcL), MT232737-MT232741(ndhF), and MW291155-MW291159 (trnL-F).
Phylogenetic analyses. We assessed evolutionary models for each region separately using jModelTest 2.1.7 (Darriba et al. 2012), and the Akaike information criterion (AIC; Akaike 1974). We analyzed each dataset separately and in combination, following a total evidence approach (Kluge 1989).
We conducted Maximum Likelihood analyses using RAxML-HPC (Stamatakis 2014) as implemented in the XSEDE tool in CIPRES (Miller et al. 2010), using a random seed (-p) of 12345, and the default (25) number of distinct rate categories (-c). For each analysis, matrices were run according to the model selected and the slow ML search algorithm. For the combined analyses including data from all three partitions (ndhF, rbcL, and trnL-F), we allowed for a mixed model (also slow search algorithm). Bootstrap analyses (100 replicates) were performed with the RAXML fast bootstrap algorithm implemented in CIPRES.
Bayesian analyses were run in MrBayes 3.2 (Ronquist et al. 2012), as implemented in CIPRES. An initial 5 million generation analysis was run using the selected model for each region to optimize parameters (including temperature) and ensure that the chains were running properly and reached stationarity. We checked chain swap information and parameter acceptance rates to ensure that parameters were acceptable (between 0.1 and 0.7), making sure all parameters had an ESS > 200, and examined appropriate chain behavior in Tracer 1.7.1 (Rambaut et al. 2018). We then conducted a second run including 20 million generations for each of our analyses. For the analyses with the concatenated dataset, the parameters associated with the model of evolution (Revmat, Statefreq, and Shape) were unlinked, while the ratemultipler, the topology and the branch lengths were linked across partitions. For all analyses, we implemented a temperature of 0.1 for the cold chain to ensure appropriate mixing. We sampled every 1,000 generations, and eliminated 25 % of the trees as burn-in. Sampling of the parameter space by the MCMC chains was summarized using the .sump and .sumt commands, while trees were visualized in FigTree 1.4.3 (Rambaut 2010).
We conducted all analyses on the Cyberinfrastructure for Phylogenetic Research cluster (CIPRES; Miller et al. 2010), which is housed at the San Diego Supercomputer Center (www.phylo.org/), and tree visualization and annotation was performed in R (R Core Team 2020).
Anatomical sampling and methods. Woods pulled from the MEXU xylarium were rehydrated in boiling water and glycerin for two hours following Pace (2019). All samples were softened in 4% ethylenediamine for two days within a paraffin oven (Carlquist 1982). Anatomical sections of the transverse, longitudinal radial and longitudinal tangential planes were performed with the aid of a sliding microtome and permanent steel knives sharpened with sandpapers of different grids (Barbosa et al. 2018). Wood sections were obtained from unembedded materials and stained in 1 % aqueous safranin. Samples with cambium and bark underwent a previous step, being gradually embedded in polyethylene glycol 1500 (Rupp 1964), and subsequently sectioned with the aid of an anti-tearing coat of a polystyrene resin (Barbosa et al. 2010). The latter were double-stained for 15 minutes in Safrablau (Bukatsch 1972, modified by Kraus & Arduin 1997). All sections were dehydrated in an ethanolic series, with butyl acetate being used in the last step, and mounted in Canada Balsam to make permanent slides.
Wood descriptions followed the IAWA Committee for hardwood (IAWA Committee 1989), IAWA Committee for bark features (Angyalossy et al. 2016), and Carlquist (2001), adjusting to the specificities of the family whenever needed. Measurements were performed using ImageJ 1.52a (National Institute of Health, USA, www.imagej.nih.gov/ij, Rasband 2012). Since approximately half of the Bignoniaceae family is composed of lianas, and it has been well-documented that lianas tend to converge to similar anatomies (Carlquist 1985, Angyalossy et al. 2012, 2015, Chery et al. 2020), we focused the comparison of Astianthus with shrub and tree members of the family.
Results
Phylogenetic placement of Astianthus. A summary of our individual and combined data matrices, including dimension, number of variable and parsimony informative characters are presented in Table 2. The GTR + gamma was recovered as the best model of DNA substitution in all analyses and implemented for all datasets.
Table 2 Diagnostics of data matrices used for phylogenetic analyses. PIC = Parsimoniously Informative Character.
| Dataset | n taxa |
n characters |
constant characters |
variable, no PIC |
PIC | frequency no PICs |
frequency PICs |
|---|---|---|---|---|---|---|---|
| ndhF | 117 | 2,176 | 1,053 | 445 | 678 | 0.205 | 0.312 |
| rbcL | 69 | 1,426 | 1,057 | 176 | 193 | 0.123 | 0.135 |
| trnL-F | 112 | 1,233 | 666 | 278 | 289 | 0.225 | 0.234 |
| concatenated | 119 | 4,835 | 2,776 | 899 | 1,160 | 0.186 | 0.240 |
The results of the Bayesian and Maximum Likelihood analyses of the combined datasets are largely congruent with those of Olmstead et al. (2009) and Lohmann (2006), including strong support for the tribes within Bignoniaceae as well as the family (Figure 2).

Figure 2 Consensus tree derived from a 20 million generation Bayesian analysis of the concatenated dataset (ndhF, rbcL, trnL-F). Both Bayesian Inference and Maximum Likelihood analyses strongly support Astianthus as sister to Campsis, within the Tecomae s.s. Posterior probabilities are provided in bold above branches and maximum likelihood bootstrap values in regular font below branches.
Our phylogenetic combined analyses based on Maximum Likelihood (ln = -38901.785924) and Bayesian Inference frameworks led almost identical topologies with minor differences not related to the placement of Astianthus, therefore only the consensus Bayesian tree is shown (Figure 2; the ML tree is available in Supplementary material 2, Figure S1). In all analyses, Astianthus is strongly supported as monophyletic (1.0 PP, 100 % ML BS). Astianthus falls within the Core Bignoniaceae clade (1.0 PP, 100 % ML BS; sensuOlmstead et al. 2009), within tribe Tecomeae s.s. (1.0 PP, 98 % ML BS), and sister to Campsis radicans (L.) Bureau (1.0 PP, 100 % ML BS).
Wood anatomy of Astianthus. Growth rings distinct, delimited by narrower vessels and radially narrow fibers (Figure 3A). Wood semi-ring porous (Figure 3A). Vessels without a specific arrangement, solitary or in radial multiples of 2-3 (Figure 3A-B), clusters of 3-4 vessels common, perforation plates simple, some wide vessel elements with foraminate perforation plate on horizontal end walls (Figure 3C). Intervessel pits alternate, minute (6 µm), vessel-ray pitting with distinct borders, similar to intervessel pits in size and shape throughout the ray cell, helical thickening absent. Vessel diameter 117 ± 32 µm, frequency 14 ± 3 vessels/mm2, two vessels per group, vessel length 258 ± 38 µm. Tyloses and deposits absent both in sapwood and hardwood. Fibers thin to thick walled (Figure 3A-C, E), with simple to minute bordered pits, septate fibers present (Figure 3E). Axial parenchyma vasicentric to aliform with short confluences (Figure 3A-B), and confluences marginating the rays (Figure 3B), with 2-4 cells per parenchyma strand (Figure 3D). Rays 3-4-seriate (Figure 3D-E), longitudinal merging of two rays common, rays lower than 1 mm. Rays either homocellular with procumbent cells only (Figure 3F) or heterocellular, with body composed of procumbent cells and one row of square marginal cells (Figure 3G). Sheath cells common (Figure 3E). Axial parenchyma cells storied (Figure 3D), and in certain areas narrow vessels also storied, but not conspicuously (storied fusiform cambial initials). Crystals absent.
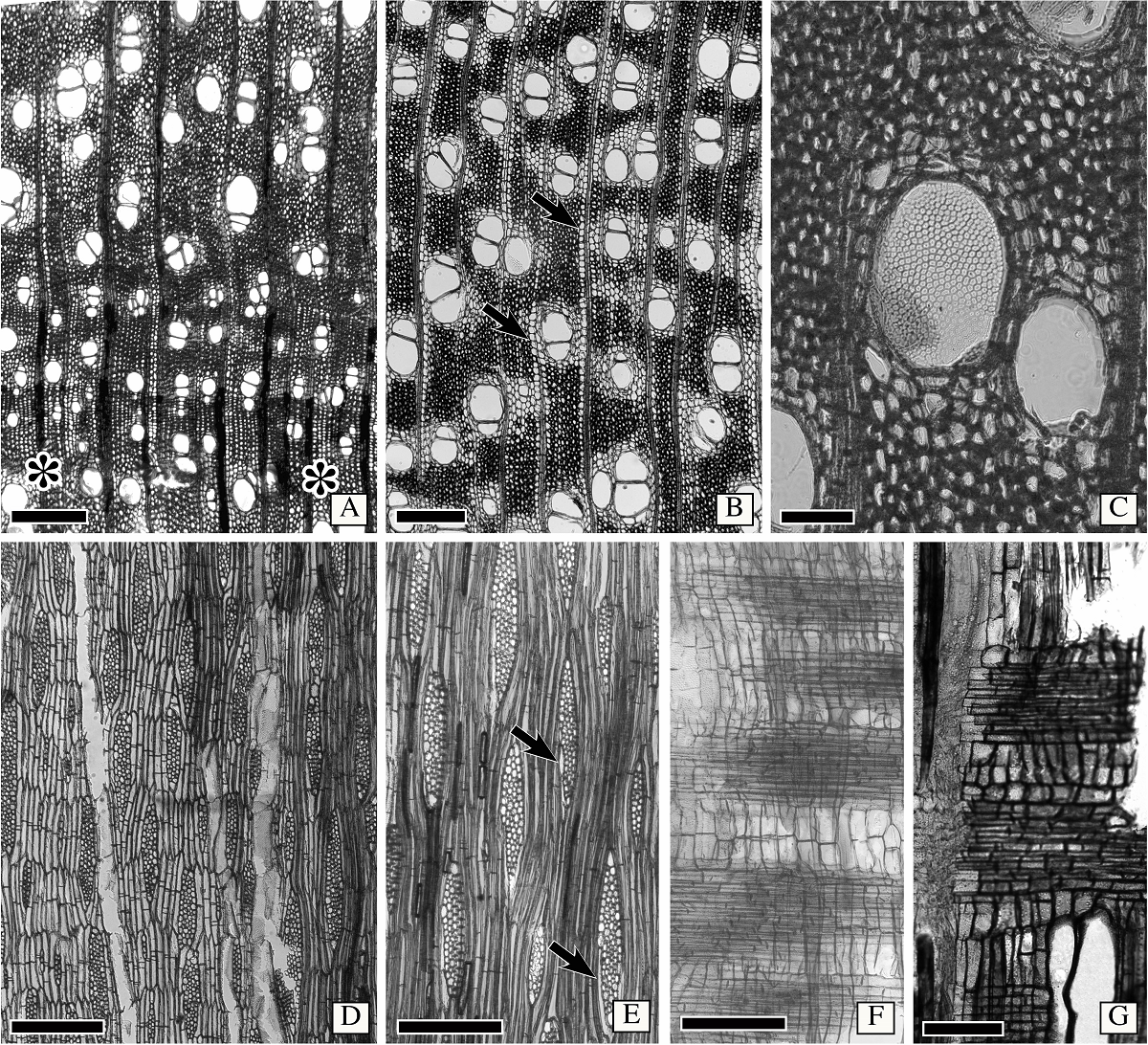
Figure 3 Wood anatomy of Astianthus. A-B. Transverse section. Semi-ring porous wood, growth rings delimited by narrower vessels, and radially narrow fibers (asterisks). Vessels solitary to multiples of 2-3. Clusters sometimes present. Fibers thin to thick walled. Axial parenchyma aliform with short confluences, some confluences also marginating the rays (arrows). C. Foraminate perforation plate in wide vessel, as seen in transverse section. D. Longitudinal tangential section. Parenchyma cells storied, with 2-(3-)cells per parenchyma strand. Rays 3-4 cells in width. E. Longitudinal tangential section. Note sheath cells present (arrows) and the septate fibers. F. Longitudinal radial section. Homocellular rays, with procumbent cells only. G. Heterocellular rays, with body composed of procumbent cells and a row of marginal square to upright cells. Scale bars: A-B, D = 400 µm, C = 100 µm, E-F = 300 µm, G = 150 µm.
Bark anatomy of Astianthus. Secondary phloem. Non-stratified phloem (Figure 4A). Conducting phloem with sieve tubes solitary or in radial multiples of 2-4 (Figure 4C). All sieve plates simple, on a transverse wall (Figure 4E). Sieve tube area 436 ± 136 µm2, diameter 24 ± 13 µm, and sieve element length of 259 ± 24 µm. One companion cell lying on the corner of the sieve tube (Figure 4C) or sometimes with two companion cells lying on opposite sides of the sieve tube (Figure 4C), companion cells in strands of more than two cells. Parenchyma constituting the ground tissue (Figure 4A, C), parenchyma strands with 2-4 cells. Course of rays straight (Figure 4A). Ray width, height and composition equal to that of the wood (Figure 4E). Ray dilatation seemingly absent (Figure 4A). Sclerenchyma composed of fibers only, diffuse, either solitary or in multiples of two (Figure 4A, C-D), with a polygonal shape (Figure 4C), differentiating close to the cambium (Figure 4C). Axial parenchyma and sieve tube elements storied. Non-conducting phloem marked by sieve tubes and companion cells empty, collapsed. Dilatation phenomena practically restricted to cell enlargement, with not much cell division in both axial and ray parenchyma. No further sclerification.

Figure 4 Secondary phloem of Astianthus and Campsis. A, C-E. Astianthus. B. Campsis. A. Secondary phloem non-stratified, diffuse fibers scattered across the entire tissue. Course of rays straight. Transverse section (TS). B. Secondary phloem non-stratified, with diffuse fibers scattered across the entire tissue. Ray course slightly undulated. TS. C. Sieve tubes in radial multiples of 2-4 common, diffuse fibers, differentiating close to the cambial region. Either one companion cell laying on one side of the sieve tube (lower arrow), or two companion cells, lying on opposite sides of the sieve tube (upper arrow). TS. D-E. Longitudinal tangential section. Fibers isolated, tapering (arrows). E. Sieve tube elements with simple, slightly inclined sieve plates (arrows). Scale bars: A-B = 300 µm, C-D = 200 µm, E = 100 µm.
Periderm. Rhytidome present, with many reticulate periderms. New periderms forming inside the secondary phloem, and enclosing large amounts of nonconducting phloem. Phellem cells evenly thin walled, non-stratified. Phelloderm cells are parenchymatous and thin walled (1-3 cell layer). No mineral inclusions recorded.
Discussion
Phylogenetic placement of Astianthus. The phylogeny of the Bignoniaceae reconstructed here indicates that Astianthus falls within Tecomeae s.s. With originally 12 genera, ca. 70 species and Pantropical distribution, Tecomeae s.s. is one of the most diverse tribes of the Bignoniaceae both in terms of morphology and distribution, with members ranging from latitudes 40o N to 40o S (Olmstead 2013) in Africa, Asia, the New World, and Oceania (Figure 5). Our analyses support Astianthus as sister to Campsis Lour., a lianescent genus with two species, one in eastern North America and one in China (Fischer et al. 2004). As currently circumscribed, tribe Tecomeae s.s. includes three main clades (Figure 5): the first clade includes the Andean herb Argylia D.Don, which is sister to the rest of the Tecomeae s.s; the second clade includes predominantly Neotropical species, with Astianthus and Campsis (except for the Chinese Campsis grandiflora (Thunb.) Schumann); the third clade consists of the rest of Tecomeae s.s., with members across the Neotropics, Africa, and Asia-Pacific (Fischer et al. 2004, Olmstead et al. 2009).
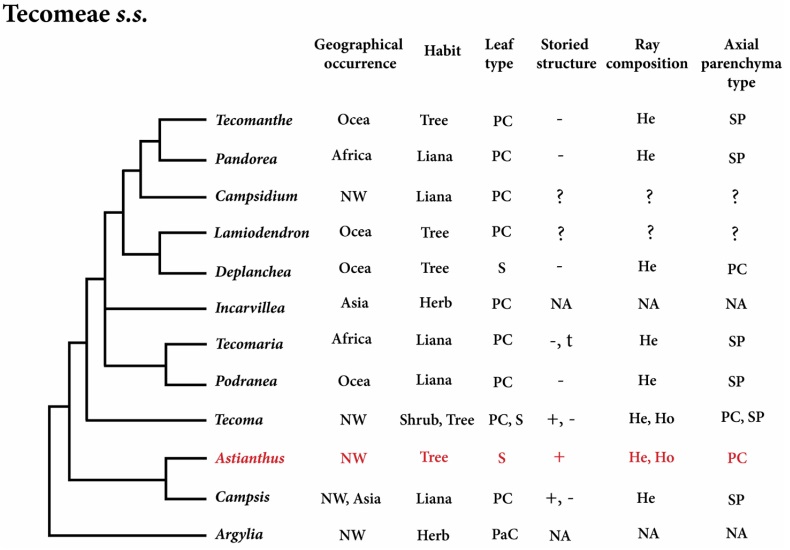
Figure 5 Phylogeny of Tecomeae s.s. with Astianthus highlighted in red. Feature comparisons. Geographical Occurrence (Africa, Asia, NW=New World, and Ocea = Oceania); Plant Habit (herb, liana, shrub or tree); Leaf Type (S = simple, PC = pinnately compound, PaC = palmately compound); Storied Structure (present or absent); Ray Composition (heterocellular and/or homocellular); Axial Parenchyma Type (PC = paratracheal confluent, SP = scanty paratracheal). NA = Not Applicable, plant without secondary growth. ? = unknown.
The placement of Astianthus within the same tribe as Tecoma corroborates Gentry’s (1992) initial proposal that the leaf similarities between Astianthus and the Catalpeae genus Chilopsis represented a convergence to their riparian habit rather than an evidence of relatedness. On the other hand, the floral and fruit similarities shared between Astianthus and Tecoma were shown to corroborate phylogenetic findinds and earlier hypotheses of Gentry (1992). Both genera share many species with yellow flowers, a cupular, 5-dentate calyx, and linear capsular fruits (Fischer et al. 2004). Tecoma is composed of 14 species of shrubs to small trees distributed in tropical America from the Andes to Arizona (Gentry 1992, Fischer et al. 2004). Most Tecoma have pinnately compound leaves, but the genus contains also some species with simple leaves, such as Tecoma castaneifolia (D. Don) Melch. from Ecuador, Tecoma tanaeciiflora (Kranzlin) Sandwith from Bolivia and Peru, and some specimens of T. weberbaueriana from Peru and Ecuador. Furthermore, nearly all species with pinnately compound leaves of Tecoma usually have simple leaves at the base of all branches (Gentry 1992).
Wood and bark anatomy of Astianthus in relation to other Bignoniaceae. Astianthus shares many wood anatomical features with other tree members of the Bignoniaceae, such as the paratracheal parenchyma with a tendency to confluences, radially thick-walled fibers delimiting growth rings, short rays, a straight grain, and rare crystals (Table 3, Figure 5; Pace & Angyalossy 2013, Pace et al. 2015a, Gerolamo & Angyalossy 2017). The presence of foraminate perforation plates in wide vessels is not found in all Bignoniaceae but is scattered in at least eight different distantly related lineages across the entire family. This feature seems to be related to species growing under strongly seasonal rain regimes (Pace & Angyalossy 2013), a hypothesis that remains to be tested.
Table 3 Synopsis of the qualitative and quantitative wood features of Astianthus and all other lineages (tribes or major clades) in Bignoniaceae.
| Tribe or clade | ASTIANTHUS | JACARANDEAE | TECOMEAE | DELOSTOMA | OROXYLEAE | CATALPEAE | BIGNONIEAE | TABEBUIA ALLIANCE |
PALEOTROPICAL CLADE |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Habit | Trees (sometimes shrubs) |
Trees, and a few subshrubs in arid zones |
Mostly lianas, with few trees and shrubs |
Trees | Trees, a few lianas | Trees | Liana, a few shrubs | Trees | Trees and shrubs | |
| Porosity | Semi - ring porous | Diffuse | Diffuse to ring - porous |
Diffuse | Diffuse | Semi - ring porous |
Diffuse to semi - ring porous |
Diffuse | Diffuse | |
| GROWTH RING MARKERS |
Marginal parenchyma |
- | + | ± | + | + | + | + | + | + |
|
Radially flattened fibers |
+ | + | + | - | + | + | + | - | ± | |
| VESSELS | Arrangement | Diffuse | Diffuse | Diffuse | Radial pattern | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse |
| Grouping | Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 & Radial multiples |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
Solitary to multiples of 2 - 3 |
|
| Vessel/group | 2 | 1.23 - 2.11 | 1.93 - 5.32 | 2.93 | 1.24 - 1.94 | 1.33 - 1.56 | 1.31 - 4.73 | 1.24 - 2.22 | 1.08 - 2.58 | |
| Dimorphism | - | - | + in lianas | - | - | - | + | - | - | |
|
Frequency (per mm2) |
14 ± 3 | 10 - 21 | 6 - 320 | 46 ± 20 | 4 - 27 | 6 - 34 | 14 - 236 | 12 - 51 | 9 - 73 | |
| Diameter (μm) | 117 ± 32 | 68 - 75 (except for J. copaia with 300) |
30 - 158 | 70 ± 12 | 80 - 179 | 131 - 204 | 45 - 293 | 44 - 125 | 51 - 178 | |
| Tyloses | - | - | - | - | - | + | - | - | - | |
| Perforation plate | Simple and foraminate |
Simple | Mostly Simple, some foraminate |
Simple | Reticulate, foraminate and simple |
Simple | Simple | Mostly Simple, some foraminate |
Mostly Simple, some foraminate |
|
|
Helical thickening |
- | - | + in species ring - porous |
- | - | + in species semi - ring porous |
- | - | - | |
|
Intervessel pit size (μm) |
6 | 7.2 - 10.3 | 4.3 - 9.4 | 3.1 | 3.1 - 5.3 | 4.1 - 11.1 | 2.6 - 12.4 | 2.5 - 19.1 | 2.2 - 10.7 | |
| AXIAL PARENCHYMA |
Patratracheal parenchyma |
Vasicentric to aliform |
Aliform | Scanty to vasicentric |
Scanty | Vasicentric to aliform |
Scanty to aliform |
Scanty to aliform |
Aliform | Aliform |
| Confluence | Short | Short to long |
Absent from present |
Absent | Short | Absent to short |
Absent to short |
Generally long, forming bands |
Short to long |
|
|
Diffuse parenchyma |
- | - | - | - | - | - | - | - | + in Coleeae | |
| Parenchyma strands | Two - four | Four (3 - 4) cells per strand |
Mostly four (3 - 4) cells per strand |
Four (3 - 4) cells per strand |
Four (3 - 4) cells per strand |
Four (3 - 4) cells per strand |
Four (3 - 4) cells per strand |
2 - 4 cells per strand |
Four (3 - 4) cells per strand |
|
| RAYS | Ray height | Short <1 mm |
Short <1 mm |
Short <1 mm and hight > 1mm in lianas |
Short <1 mm |
Short <1 mm |
Short <1 mm |
Generally high >1 mm, smaller in shrubs |
Short <1 mm |
Short <1 mm |
|
Ray width (in number of cells) |
4 - Mar | 1/2 - 3 | 2 - 3 | 3 | 3 | 3 | 1 - 9 | 1 - 3 | 1 - 3 | |
|
Rays: cellular composition |
Mostly homocellular, some heterocellular |
Homocellular in Jacaranda Monolobos and heterocellular in Jacaranda Dilobos |
Heterocellular | Homo and hetero with 1 row of square cells |
Homocellular | Homo and hetero with 1 row of square cells |
Heterocellular mixed | Homocellular | Homo and hetero with 1 row of square cells |
|
|
Vessel - ray pitting |
Similar to intervessel pits |
Similar to intervessel pits |
Similar to intervessel pits |
Similar to intervessel pits |
Similar to intervessel pits |
Simple to semi - bordered |
Predominantly similar to intervessel pits |
Similar to intervessel pits |
Similar to intervessel pits |
|
|
Perforated ray cells |
- | - | + in lianas | - | - | - | + | - | - | |
|
Septate fibers |
+ | - | + | + | ± | ± | + | - | - | |
|
Storied structure |
+ | - | - | - | - | - | - , present in but a few species |
+ | - | |
| Crystals | - | Present in the rays of some species |
Present in the rays of some species |
Present in rays | - | Present in the rays of some species |
Present in the rays of some species |
When present, in both rays and axial parenchyma |
Present in the rays of some species |
|
Considering less common anatomical attributes, Astianthus would still be a good fit in at least three different Bignoniaceae major clades: the Paleotropical clade, the Tabebuia alliance, and Tecomeae s.s. (Table 3). However, based on the Neotropical distribution, Astianthus is best placed in the Tabebuia alliance or Tecomeae s.s.. Members of both tribes can have a storied structure (although this feature is more common in the Tabebuia alliance) and homo to heterocellular rays (Pace et al. 2015a). However, the combination of these two features in addition to the presence of septate fibers is found exclusively in Tecomeae s.s. (Table 3), supporting the phylogenetic placement suggested by the molecular data. The most notable differences between Astianthus and some members of Tecomeae s.s. (Figure 5) are likely associated to the difference of habits. Many of the genera of Tecomeae s.s. include lianas that seem to converge in a reduction in the wood axial parenchyma (Pace & Angyalossy 2013), contrary to what is found in lianas from other plant families (Angyalossy et al. 2015). In addition, in lianas in general the rays tend to become more heterocellular, similarly to what was seen in other Bignoniaceae, especially in the lianescent tribe Bignonieae (Pace & Angyalossy 2013).
The morphological similarity between Astianthus and members of the North American tribe Catalpeae is not mirrored by the wood anatomy. Members of Catalpeae are marked by a heartwood with abundant tyloses, a non-storied structure, and vessel to ray pits simple to slightly bordered (Pace et al. 2015a). On the other hand, Astianthus lacks tyloses, has storied axial parenchyma, and distinctly bordered vessel to ray pits.
The bark anatomy provides further support for the inclusion of Astianthus in Tecomeae s.s. Virtually all Bignoniaceae species studied thus far show a stratified bark, with clear fiber bands alternating with axial parenchyma and sieve tubes, regardless of the habit, ecological factors, or distribution (Roth 1981, Pace et al. 2011, 2015b). The single exception to this rule is Campsis, which emerged as sister to Astianthus, with whom it shares scattered single fibers across the entire phloem (Evert 2006, Figure 4B), a potential synapomorphy of this clade. This finding corroborates previous assumptions that the bark anatomy carries a strong phylogenetic signal in the family, independently of the habit, aiding the delimitation of major clades within the family (Pace et al. 2015b). Other bark features of Astianthus such as the presence of sieve tubes in radial multiples, axial parenchyma as a background tissue, a seemingly absent ray dilatation by cell divisions, and a reticulate rhytidome are more widespread in the family (Roth 1981, Pace et al. 2015b).
In conclusion, our phylogeny reconstruction based on three plastid markers (ndhF, rbcL, and trnL-F) indicates that Astianthus is nested within Tecomeae s.s.. This placement is further supported by the non-stratified bark, scattered bark fibers, storied axial parenchyma, homo and heterocellular rays co-occurring, and septate wood fibers. These results show the importance of combining in-depth studies of morphology and anatomy with molecular phylogenetic data for an improved understanding on plant diversification, especially in the tropics. The placement of Astianthus within Tecomeae s.s. further supports a neotropical origin for the tribe.
Supplementary material
Supplemental data for this article can be accessed here: https://doi.org/10.17129/botsci.2779











 nueva página del texto (beta)
nueva página del texto (beta)



