The coccoid green algae are one of the most troublesome taxonomic groups (Eliáš et al. 2010). Scenedesmaceae is the largest family with 54 genera (Guiry & Guiry 2016), each one displaying great morphological variability, maintained by genetic relationships resulting from autospores (Hegewald 1997), that consequently promote all other mutations (Krienitz & Bock 2012).
Simple morphology, phenotypic plasticity and the presence of cryptic species in the family Scenedesmaceae have contributed to taxonomic complication (Sciuto et al. 2015), resulting in constant reassignments (Eliáš et al. 2010), such as that which has occurred with various species of the Coelastrella subfamily Coelastraceae (Hegewald et al. 2010). However, the body of molecular data has supported the transfer of those specimens to other genera; e.g., Coelastrella multistriata possesses a close relationship with Scenedesmus vacuolatus. Despite morphologic differences, genetic similarities based on 18SrDNA supported the transfer of Coelastrella multistriata into Scenedesmus (Hanagata 1998). This is because the Coelastraceae subfamily with 11 species possesses a close relationship with the Scenedesmaceae family, despite morphologic differences. Coelastraceae show spherical, elliptical and long coenobia (Punčochářová & Kalina 1981), while Scenedesmaceae show flat, straight, and lightly curved coenobia (Guiry & Guiry 2016). Nevertheless, the close genetic relationship suggests that the subfamily Coelastroideae should be treated as the genus Coelastrella within Scenedesmaceae together with Coelastrum, Hariotina, Asterarcys and Dimorphococcus (Hegewald et al. 2010).
Coelastrella, named by Chodat (1922), shows distribution in Korean rivers (Song & Singh 2014), Australian soil (Tschaikner et al. 2007, Tschaikner et al. 2008), rock surfaces (Abe et al. 2004), Bulgarian soil (Uzonov et al. 2008), and an alpine zone in New Zealand (Gopalakrishnan et al. 2014). It was recently reported for the first time in freshwater in North America (Neofotis et al. 2016), but prior to this study it had not been reported in a tropical region of North America (Yucatan Peninsula). Its habitats range from aerial to terrestrial with relative humidity (40 %) or without (Song & Singh 2014), as well as at high altitudes (Gopalakrishnan et al. 2014).
Coelastrella shows morphologic traits similar to Scotiellopsis (Punčochářová & Kalina 1981). Although both belong to the Coelastroideae subfamily (Guiry & Guiry 2016), they are characterized by spherical to elliptical coenobia with meridional ribs on the cell surface, but they differ in number and polar thickening (Hanagata 2001). These structures are generally not visible under light microscopy, because there are also coenobia that lack cell wall ridges (Hegewald et al. 2010). In contrast, electron microscopy has aided elucidation based on these structures, although sometimes the cell wall might be artificially wrinkled during sample preparation (Kaufnerová & Eliáš 2013). Cell wall sculptures observed in Coelastrella by electron microscopy can be helpful for their identification (Tschaikner et al. 2007) meanwhile molecular identification will become a universal tool in botanical studies (Manoylov 2014). In this regard, ITS-2 sequencing has been successfully used to verify the taxonomy of closely related species (Ruhl et al. 2010), while ITS1-5.8S-ITS2 has been used to successfully confirm the identification of freshwater green microalgae strains (Timmins et al. 2009, Hadi et al. 2016). The ITS1-5.8S-ITS2 region from virtually all Viridiplantae can be amplified with a single set of universal primers (White et al. 1990), despite these markers being of high variability (Hall et al. 2010). The delimitation in Coelastrella needs more attention (Kaufnerová & Eliáš 2013). In fact, it has been suggested that morphological diversity raises the question of whether these algae are a natural assemblage or an artificial group (Hegewald et al. 2010). In this study, three strains from shallow waters of Yucatan were examined using optical and electron microscopy for ultrastructure analysis and using molecular data based on ITS-2 and ITS1-5.8S-ITS2 phylogenetic studies to gain more insights into their taxonomic status.
Material and methods
Microalgal strains. Three strains labeled CORE-1, CORE-2 and CORE-3 were studied. All of them were provided by the Marine and Freshwater Microalgae Collection of the Yucatan Peninsula (FICOYUC) at the Alfredo Barrera Marin Herbarium of the Universidad Autónoma de Yucatán in Mérida, Yucatán, México. All microalgae were first maintained in Bristol medium at 27 °C, 12:12 light-dark, with a light intensity of 100 µmol/m2/s. Growth curves for each microalga were carried out in 50 ml Erlenmeyer flasks containing 10 ml fresh TAP medium (Gorman & Levine, 1965). They were incubated at 27 ± 2 °C, 120 rpm (Newbrunswick Scientific, Excella E10), and a light intensity of 100 µmol/m2/s with a 16:8 (light/dark) photoperiod. Samples were taken every day (500 µL) and cell concentration was determined using a hemocytometer (Andersen 2005).
Morphological analysis. Morphology features observed through optical microscopy (Nikon, Eclipse E200) were recorded, placing special emphasis on cell wall structures as a means of taxonomic identification by SEM micrographs. Lugol’s solution (Jalmek SY050-13) was used for pyrenoid staining with the following procedure: 50 µL of Lugol’s solution was mixed in 200 µL of microalgae culture. Subsequently, one drop (10 µL) was taken and visualized under light microscopy. Starch granules were observed from pyrenoid staining (Kumar & Singh 1971, Throndsen 1978).
Scanning electron microscopy (SEM). For the SEM analysis, 200 mg of microalgal cells in stationary phase were used, because the growth stops and almost the same cell size is observable in all cells (Berge et al. 2012). They were recovered and centrifuged (Hettich, Mikro 200R) at 9,727 ×g for 20 min. The supernatant was discarded and the pellet was transferred to a nylon membrane (GE Healthcare Biosciences, RPN203B) and kept in desiccators under vacuum for 24 h in 2.5 % glutaraldehyde (Polysciences, 00376) at 4 °C. This procedure was repeated three times to guarantee cell fixation. Fixation agent was then removed by centrifugation at 9,727 ×g for 20 min. The pellet was subsequently washed six times in 0.2 M phosphate buffers solution (pH 7.1). Sequential dehydration was performed in an ethanol series (30, 40, 55, 70, 85 and 100 %) at 4 °C. Individual samples were placed in microporous specimen capsules (C1178, Sigma) to be dried using Critical Point Dryer (Samdri 795). The samples were then mounted on specimen mounts for scanning electron microscopes and gold coated (Desk-II Cold sputter/ETCH Unit). SEM was performed (JEOL JSM6360 LV) (Kaufnerová & Eliá? 2013).
Genetic identification. At the stationary growth phase, the microalgae cells were harvested by centrifugation (OrtoAlresa, Digicen 21R) (30 ml) and washed three times in bidistilled sterile water. The cells (c.a. 200 mg) were stored at -20 °C, until use. For DNA extraction, we followed the protocol modifications of Youssef et al. (2015).
ITS-2 and ITS1-5.8S-ITS2 amplification. ITS-2 and ITS1-5.8S-ITS2 amplifications were implemented using the forward and reverse primers proposed by Van Hannen et al. (2002) and Timmins et al. (2009) respectively. Both assays were carried out with 10 µL of reaction mixture containing 1x PCR Buffer, 2 mM MgCl2, 0.2 mM dNTPs (Bioline, DM109H), 1 mM primer, 1 U Taq DNA polymerase recombinant (Invitrogen 11,615-010) and 10 ng of DNA. PCR conditions for ITS-2 included 1 cycle of 1 min at 94 ºC for initial denaturation, followed by 35 cycles of 1 min at 94 ºC, 1 min at 55 ºC, 1.5 min at 72 ºC, and finally 7 min at 72 ºC. Conditions for ITS1-5.8S-ITS2 amplifications were 1 cycle of 5 min at 95 ºC for initial denaturation, followed by 30 cycles of 40 s at 95 ºC, 40 s at 56 ºC, 40 s at 72 ºC, and finally 10 min at 72 ºC. Amplicons were checked by electrophoresis on 1 % agarose gel in 1 x TAE buffer. The amplification products (fragments of approximately 314 pb from ITS-2 and 714 bp from ITS1-5.8S-ITS2), were purified using NucleoSpin® Gel and the PCR Clean-up commercial kit (Macherey-Nagel, Düren, Germany) to eliminate primers dimers according to the manufacturer’s instructions. Direct sequencing of the amplification products was carried out by a two-directional (reverse and forward) procedure (Clemson Sequencing Service).
The PCR products from ITS-2 sequencing showed double peaks in the electropherogram generated as an output of direct sequencing. They were therefore inserted in pGEM-T Easy Vector System (A1360, Promega) and cloned in Escherichia coli (DH5α) cells. The plasmids were purified with QIAprep Spin Miniprep Kit (27104, QIAGEN). The sequencing was subject to both forward and reverse sides using M13 primer.
ITS-2 and ITS1-5.8S-ITS2 sequence analysis. The ITS-2 (314 pb) and ITS1-5.8S-ITS2 sequences (714pb: 27pb of 18S rDNA; 213pb of ITS-1; 181pb of 5.8S rDNA and 280 pb of ITS-2) were subjected to a BLAST search to determine their identities and assess their homologies and similarities to those in GenBank. Sequences with E-value closer to 0.00, 100 % coverage and identity 96 % > with each entry were aligned using CLUSTALX (EMBL, Heidelberg, Germany) and manually adjusted by MEGA version 2.1. Molecular phylogenetic analyses by the maximum likelihood method based on the Kimura 2-parameter model (Kimura 1980) were conducted using MEGA version 2.1 (Kumar et al. 2016). One thousand replica samplings were analyzed for percent bootstrap values in a neighbor-joining tree. The percentage of trees in which the associated taxa clustered together is shown next to the branches. A discrete Gamma distribution was used to model evolutionary rate differences among sites (2 categories (+G, parameter = 0.0500)). Evolutionary analyses were conducted in MEGA 7 (Kumar et al. 2016). Reference sequences retrieved from GenBank of the ITS1-5.8S-ITS2 sequences were KX940913, KX940914, KX940915 for CORE-1, CORE-2 and CORE-3 respectively.
Results
Morphological analysis. The morphology observed by light microscopy (LM) was complemented by scanning electron microscopy (SEM) micrographs. CORE-1 showed an ovoid to ellipsoidal cell shape and rough cell wall. Asexual reproduction was visible by autosporulation. Each sporangium was composed of 2-6 young autospores with visible pyrenoid (Figure 1 C). The cell arrangement in sporulation showed cells in two superposed lines. The cell measurements were 7.01±0.28 µm in length and 4.56±0.31 µm in width with some granulations of 159 nm dispersed on the cell wall (Figure 2 A-B: indicated by arrows). The pyrenoid was defined by two or three divisions of starch granules.

Figure 1 Vegetative cells of microalgae strains. A-C. CORE-1. A. unicellular organization of the cells. B. pyrenoid visible. C. autospores with pyrenoids. D-F. CORE-2. D. colonial organization. E. pyrenoid staining with starch cells. F. autospores in sporangium indicated by arrows. G-I. CORE-3. G. colonial organization. H. Visible starch sheaths of the pyrenoid. I. autospores in sporangium. J. orange appearance of the cells.

Figure 2 A-F. SEM of the cells of the study. CORE-1. A. cell wall appearance (granulations indicated by rows). B. well pronounced apex. CORE-2. C. vegetative cells. D. smooth appearance of the cell. CORE-3. E. meridional ribs. F. forked and well pronounced apex.
CORE- 2 showed spherical and ellipsoidal cell shapes with one pole lightly developed (Figure 1 D-F). The cell was of 8.4±1.2 µm in length by 6.5±0.91 µm in width (Figure 2 C-D). The cell wall was smooth and 2-8 autospores were observed per sporangium (Figure 1 F: indicated by arrows). The pyrenoid was observed after Lugol staining, showing mostly continuous starch granules.
CORE-3 showed ovoid cells of 10.0 ± 0.23 µm in length and 6.7 ± 0.14 µm in width. A smooth cell wall was observed via LM (Figure 1; G-I), while ribs in a meridional distribution were observed via SEM (Figure 2; E-F). One of the ends was observed to be forked and well pronounced. Autospore formation (2-8 autospores) was observed. A visible pyrenoid composed of two or three divisions of starch plates was observed, sometimes observed to be continuous without divisions and with the vacuole visible. One interesting characteristic was the orange coloration of cells in older cultures (Figure 1; J).
Genetic analysis. The sequences of each of the three strains were used as a “query” in nucleotide Blast tools (NCBI website) to recover the ten best matches for each input sequence, based on percentage of sequence coverage, E-value and percentage of identity. The sequences selected were used for ITS-2 and ITS1-5.8S-ITS2 rDNA phylogenetic tree construction. These displayed clusters with bootstrap values above 50 %. The length of each branch denoted evolutionary distance.
The phylogenetic tree resulting from ITS-2 sequences (Figure 3) presented a high support. CORE-1 was clustered with good bootstrap support, near to Asterarcys sp. MS3 and Coelastrella sp. M60. CORE-2 was well supported in its monophyly with Coelastrella sp. accessions (KU507, KU502). Meanwhile, CORE-3 was clustered with high support, but evolutionarily separate from other strains, and in a node dominated by the genus Coelastrella, together with Chlamydomonas moewusii and other related species, such as Haematococcus pluvialis and Imbribryum alpinum.

Figure 3 Phylogenetic tree inferred from ITS2 rDNA sequences constructed with the neighbor-joining method. The number of the branches indicated bootstrap values (1,000 replicates).
Using ITS1-5.8S-ITS2 sequences, the phylogenetic relationships were very similar and resolved at 50 % bootstrap values (Figure 4). CORE-1 forms a node with Scenedesmus rotundus CFR 1-06/FW, Asterarcys sp. MS3, and Coelastrella sp. CORE-2 was placed close to the same Coelastrella accessions, as in the ITS-2 phylogenetic tree. Meanwhile CORE-3 was paraphyletic with high support and clustered with others accessions of Coelastrella genera. This clustering confirms in a certain way, the relationship of all strains with Coelastrella species. It is important to mention that the phylogenetic tree constructed by both ribosomal regions analyzed showed well supported nodes. Therefore, the genetic relationship of the strains under study must be considered for taxonomic assignment.
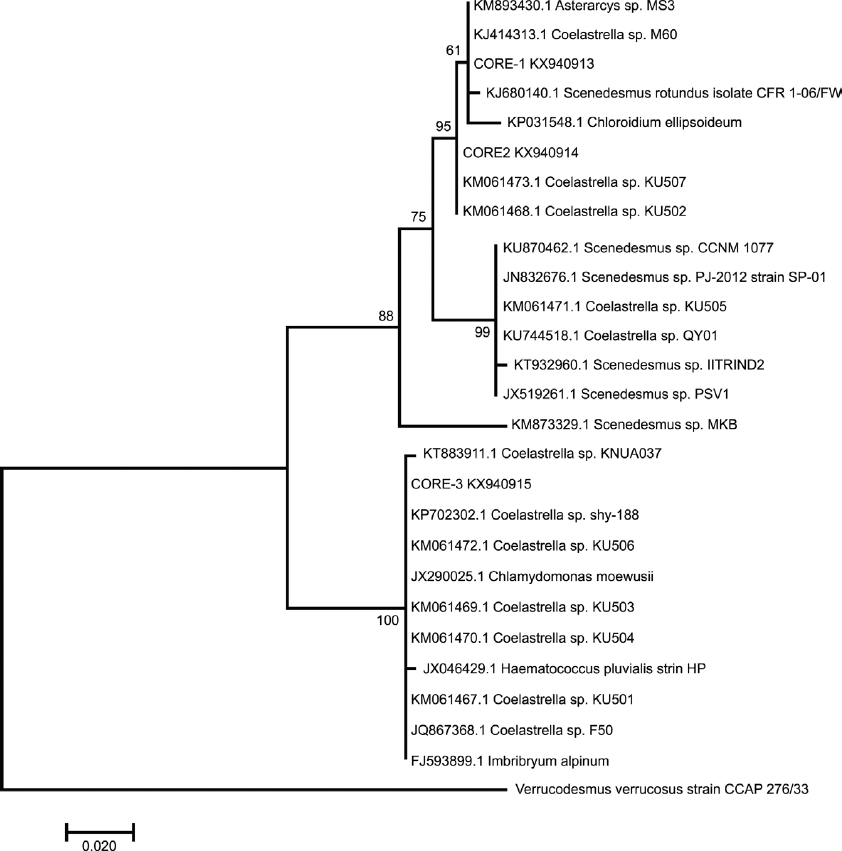
Figure 4 Phylogenetic tree inferred from ITS1-5.8S-ITS2 rDNA sequences constructed with the neighbor-joining method. The number of the branches indicated bootstrap values (1,000 replicates).
Verrucodesmus verrucosus was included to contrast taxonomically with CORE-1, but it was clustered as an external outgroup in both phylogenetic trees.
Discussion
Morphologic analysis indicated that all strains belong to the family Scenedesmaceae. The autospore formation characteristic of Scenedesmaceae was visible in all strains under study, observed on light microscopy (Figure 1). SEM analysis of CORE-1 showed visible granulations dispersed over the cell wall surface of some cells and scarce in others (Figure 2; A-B). The granulations are a taxonomic character with identity to Scenedesmus verrucosus (Y.V. Roll) (Hegewald et al. 2013, Ramos et al. 2015), a basionym of Verrucodesmus verrucosus (Y.V. Roll) from their separation by rRNA gene sequences (ITS1/5.8S/ITS2) (Hegewald et al. 2013). Two lines of 6 intercalated cells were observed (Figure 1; C). Verrucodesmus verrucosus exhibits the arrangement of cells in two series of 4 or 8 cells as a taxonomic trait (Ramos et al. 2015). Thus, based on morphological analysis, CORE-1 belongs to V. verrucosus.
CORE-2 is rather simple and does not show any structure on the cell wall that could aid with its identification.
In CORE-3, meridional ribs were lightly evident, corresponding to the genus Coelastrella (Chodat 1922). Coelastrella belongs to the Scenedesmaceae (Hegewald & Hanagata 1999). This species is aerial to terrestrial with humidity (40 %) or without (Song & Singh 2014), and at high altitudes (Gopalakrishnan et al. 2014). It shows distribution in different regions (Abe et al. 2004, Tschaikner et al. 2007, Tschaikner et al. 2008, Uzonov et al. 2008, Song & Singh 2014), and was recently reported in North America (Neofotis et al. 2016). However prior to this study Coelastrella had not been reported in tropical regions of North America (Yucatan Peninsula).
CORE-3 showed meridional ribs characteristic of Coelastrella (Uzonov et al. 2008), and should therefore be considered Coelastrella sp. In CORE-3, an orange coloration was also observed in older cultures under light microscopy, consistent with Coelastrella specimens, which show a red (Guiry &Guiry 2016) or orange coloration (Hu et al. 2013) in matured cultures, similar to that of Coelastrella striolata var. multistriata (Trenkwalder) dependent on a low nitrogen concentration in the culture medium (Abe et al. 2004). The autospore number of CORE-3 (2-8) was different to Coelastrella members (Chodat 1922), which show 2-18 autospores released from mother cells (Guiry & Guiry 2016).
The rDNA sequences analyzed provided more insights into the taxonomic status. This is probably due to the increased number of taxa verified in algal gene libraries (Manoylov 2014). Phylogenetic analysis of the genus Coelastrella possesses higher molecular diversity, as suggested by our results via both ITS-2 and ITS1-5.8S-ITS2 sequence analysis and those of Kaufnerová and Eliáš (2013). Interestingly, through the ITS-2 or ITS1-5.8S-ITS2 phylogenetic trees, the positions of clades in the tree were resolved based on the high support.
The ITS-2 sequenced region has helped to discriminate at the species level, and clarified the relationships of Coelastrella with other genera of the family Scenedesmaceae, such as Hariotina, Asterarcys, Coelastrum and Dimorphococcus (Hegewald et al. 2010).
Contrary to the morphological analysis that showed identity with V. verrucosus, CORE-1 was nested within Coelastrella and Asterarcys by ITS-2 sequences and with the genera Coelastrella and Scenedesmus by ITS1-5.8S-ITS2 sequences, consistent with clustering by 18S rDNA sequences (Kaufnerová & Eliáš 2013) and ITS-2 region (Hewegald et al. 2010). Initially the Coelastraceae subfamily includes Asterarcys, a very different morphologic genus but one which has been nested between taxa nominally added to Coelastrella. A similar situation occurs with Scenedesmus rotundus (Hegewald et al. 2013) or Scenedesmus rubescens, which maintain close relationships with the genus Coelastrella (Chodat 1922), probably due to minor differences in the 18S rDNA sequence (Kaufnerová & Eliáš 2013), which were evidenced by the genetic markers analyzed in this study.
Morphology observed in CORE-1 indicated identity with Verrucodesmus verrucosus. However, molecular traits from two taxonomic markers indicated no genetic relation with V. verrucosus. Its position in the phylogenetic tree is very far away, and it can even be considered an external outgroup. Thus, it is not possible to conclude that CORE-1 belongs to V. verrucosus. On the contrary, there is a strong possibility that it is genetically closest to Coelastrella and distant to V. verrucosus.
CORE-2 forms a monophyletic clade with Coelastrella accessions reported as hydrogen producers (Pongpadung et al. 2015). Considering the absence of structures on the cell wall, it was not possible suggest taxonomic identity. However, based on molecular analysis CORE-2 should be considered to belong to Coelastrella.
The taxonomic identity of CORE-1 and CORE-2 was based on phylogenetic evidence. This has already been performed for members of Coelastrella, e.g. Coelastrella multistriata possesses a close relationship with Scenedesmus vacuolatus. Despite morphologic differences, genetic similarities based on 18SrDNA supported transfer of Coelastrella multistriata into Scenedesmus (Hanagata 1998).
CORE-3 formed one clade with Coelastrella and other very different genera with low support: Haematococcus pluvialis, Chlamydomonas moewusii (CCAP 11/64A) and Imbribryum alpinum. The only relation between CORE-3 and Haematococcus is the coloration of cultures due to astaxanthin production capability, because this genus is not related to the family Scenedesmaceae. On the other hand, it has been suggested that Chlamydomonas moewusii (CCAP 11/64A) was misidentified because it is morphologically different from Coelastrella (Kaufnerová and Eliáš, 2013). Imbribryum alpinum is also very different from the other species. The morphology and molecular characters of CORE-3 were consistent with Coelastrella, and the genetic results indicate that it probably belongs to Coelastrella.
Based on the results obtained, it is possible to suggest that Coelastrella members do not always show morphological traits congruent with Coelastrella descriptions. This is probably due to high susceptibly to phenotypic plasticity, which interferes with taxonomic assignments based only in morphology. The sequencing of molecular markers employed in this study was used for taxonomic assignment, supporting the assertion by Manoylov (2014) that “molecular identification will become a universal tool in biological studies”. In this regard, this report contributes to knowledge of Coelastrella species and provides the first description of three of them found in a tropical region of North America in the Yucatan Peninsula.
Conclusions
The three strains under study showed morphologic characteristics of the family Scenedesmaceae. The morphologic and molecular analysis of CORE-3 was close to the taxonomic descriptions of Coelastrella. Morphologic and genetic analysis of CORE-1 and CORE-2 rendered different results. While the morphologic traits of CORE-1 were quite similar to Verrucodesmus verrucosus, genetic analysis indicated a relationship with Coelastrella species, and did not support morphologic identity with V. verrucosus. CORE-2 did not show taxonomic traits because of the absence of structures. However, genetic analysis indicated that this strain belongs to Coelastrella.
Therefore, based on the high support of the nodes of the phylogenetic analysis by ITS-2 and ITS1-5.8S-ITS-2 sequences, all strains under study belong to the genus Coelastrella. Further sequencing analysis with 18S rDNA could contribute to supporting the phylogenetic status of CORE-1 and CORE-2 within Coelastrella.











 nueva página del texto (beta)
nueva página del texto (beta)


