Introduction
Diarrhea continues to be an important health issue worldwide (Thakur et al., 2018), particularly in developing countries and children younger than 5 years old and in the elderly. Its etiology can be viral, bacterial or parasitic. Viruses represent 70 to 90 % of diarrhea cases in children; and bacteria from 10 to 20 %. such as Salmonella, Shigella, Campylobacter, enterotoxigenic Escherichia coli and less frequent enteroinvasive Escherichia coli. With respect to parasites Giardia lamblia, Entamoeba histolytica and Cryptosporidium spp represent less than 5 % of cases (Florez, 2020). Among the bacteria associated with diarrhea there are different Escherichia coli (E. coli) pathotypes, which colonize the human intestine, can directly spread from person to person, from animal to person or indirectly through water or contaminated food (Sperandio & Nguyen, 2012). According to its pathogenesis and epidemiologic characteristics, this group of bacteria is classified into six pathotypes: enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive (EIEC), enteroaggregative (EAEC), diffusely adherent (DAEC) and enterohemorrhagic (EHEC), Shiga toxin-producing (STEC), as well as toxin producer Verocytotoxin-producing (VTEC) (Kaper et al., 2004). The Center for Disease Control and Prevention of the United States (CDC) published in its morbidity and mortality weekly report that ETEC has been related with diverse outbreaks associated to foods and water in developed and developing countries (CDC, 1994; FDA, 2011). Since O157:H7 E. coli is frequently found in stool of cattle, this is one of the main sources of contamination toward beef and its derivatives; due to sanitary deficiencies during working, slaughter, and eviscerating for obtaining the carcass (Stanley et al., 2017). Consumption of poorly cooked and contaminated ground beef with EHEC has been a cause of bloody diarrhea that can evolve into hemolytic-uremic syndrome (HUS) (Surendran-Nair, 2017). Beef carcass and its derived cuts have been reported as the source of infection in many outbreaks caused by EHEC (Barham et al., 2002). The first outbreak documented in the United States of America (USA) was in 1982, which was associated to ground beef, and up to day, it remains as the most common vehicle among disease outbreaks caused by E. coli pathotypes (Ranger et al., 2005). 0157:H7 serotype is the main cause of outbreaks associated to E. coli in USA (Karmali et al., 2010).
Bovine meat has been observed to be implicated into outbreaks by EHEC worldwide and in Mexico it is a staple food (Kiranmayi et al., 2010; INEGI, 2018), reason why it is important to demonstrate the potential presence of the different E. coli pathotypes in food. In this context, the aim of the work was to isolate and identify E. coli pathotypes in ground beef, sold in butcher’s shops in Guadalajara, Jalisco.
Material and Methods
Samples collecting
For this descriptive study of prevalence, total of 100 samples of ground beef were collected, 100 from “traditional” butcher’s shops (10 per month) in Guadalajara, Jalisco. The sampling period was comprised from February to November 2014 (Table 1), warm months for this region, in order to increase the possibility of isolation according to recommendations of the study of Varela-Hernández et al. (2007) and Cagney et al. (2004). A sample bigger than 50 g was collected in a sampling bag (Speci-Sponge, Nasco Whirl-Pak®, Modesto, Ca, USA) and transported in containers with refrigerant gel (4-8 ºC), to the Laboratory of Medical Sciences of the Ciénega University Center for its analysis in a time no longer than 24 h.
Microbiologic analysis
For this study, a sample of 25 g of fresh ground beef was taken, to which 255 mL of diluent were added, prepared with casein peptone (BD®, Bioxon®, Becton Dickinson®, México) and 0.85 % NaCl (Sigma-Aldrich®, México). Afterwards, the sample was homogenized by using a homogenizer (Stomacher® Biomaster 80, Port Saint Lucie, FL, USA), at a velocity of 60 rpm, for two minutes. For the isolation of suspect strains of pathogenic E. coli (EIEC, EPEC, ETEC, DAEC AND EAEC), the method proposed by the FDA (2011) was used, where the isolations were realized in eosin methylene blue agar (EMB agar) (bluishblack colonies with metallic green shine) and MacConkey agar (large colonies, intense pink with precipitation halo). For enterohemorrhagic E. coli, the protocol proposed by Varela et al. (2007) was followed, in which isolation was performed using immunomagnetic separation with Dynabeads® (Dynal Biotech, Norway), immunomagnetic beads were inoculated in MacConkey agar with sorbitol supplemented with Cefixime and Tellurite (colorless or beige colonies) and CHROMagar® agar (mauve colonies). Biochemical tests for the isolated strains were realized using the API20E system (bioMérieux, France). Control pathotypes used in this study were shown on Table 2.
Table 2 Escherichia coli strains used as a positive control to standardize amplification protocols.
| Pathotype | Amplified virulence factor | Serotype or identification code | Origin* |
|---|---|---|---|
| EHEC | stx1, stx2, eaeA, hly933 | O157:H7 | ATCC 46597 |
| EHEC | stx1, stx2, eaeA, hly933 | O157:H7 | ATCC 43895 |
| EPEC | eaeA, hly933 | O111 | ATCC 43887 |
| ETEC | est, elt | E9034A | Universidad Autónoma de Puebla |
| EPEC | eaeA, hly933 | E2348169 | Universidad Autónoma de Puebla |
| EAEC | aggR | O42 | Universidad Autónoma de Puebla |
| EIEC | ipaH | O28ac:H5 | Universidad Autónoma de Puebla |
*Collection of the indicated institution.
Determining pathotypes by PCR
The strains obtained by both methods of isolation and of a control strain of each pathotype were submitted to DNA extraction, using the protocol proposed by Feng & Monday (2000). The quality of the DNA was assessed through observation in an 1 % agarose gel, stained with ethidium bromide and the concentration was obtained through spectrophotometry. Likewise, the DNA/proteins ratio was determined, to assess its quality. Calibration curves of magnesium chloride were carried out for each of the three polymerase chain reactions (PCR) used in this study, using DNA from the control strains (Figure 1). The conditions and optimal components of each of them were shown on Table 3.
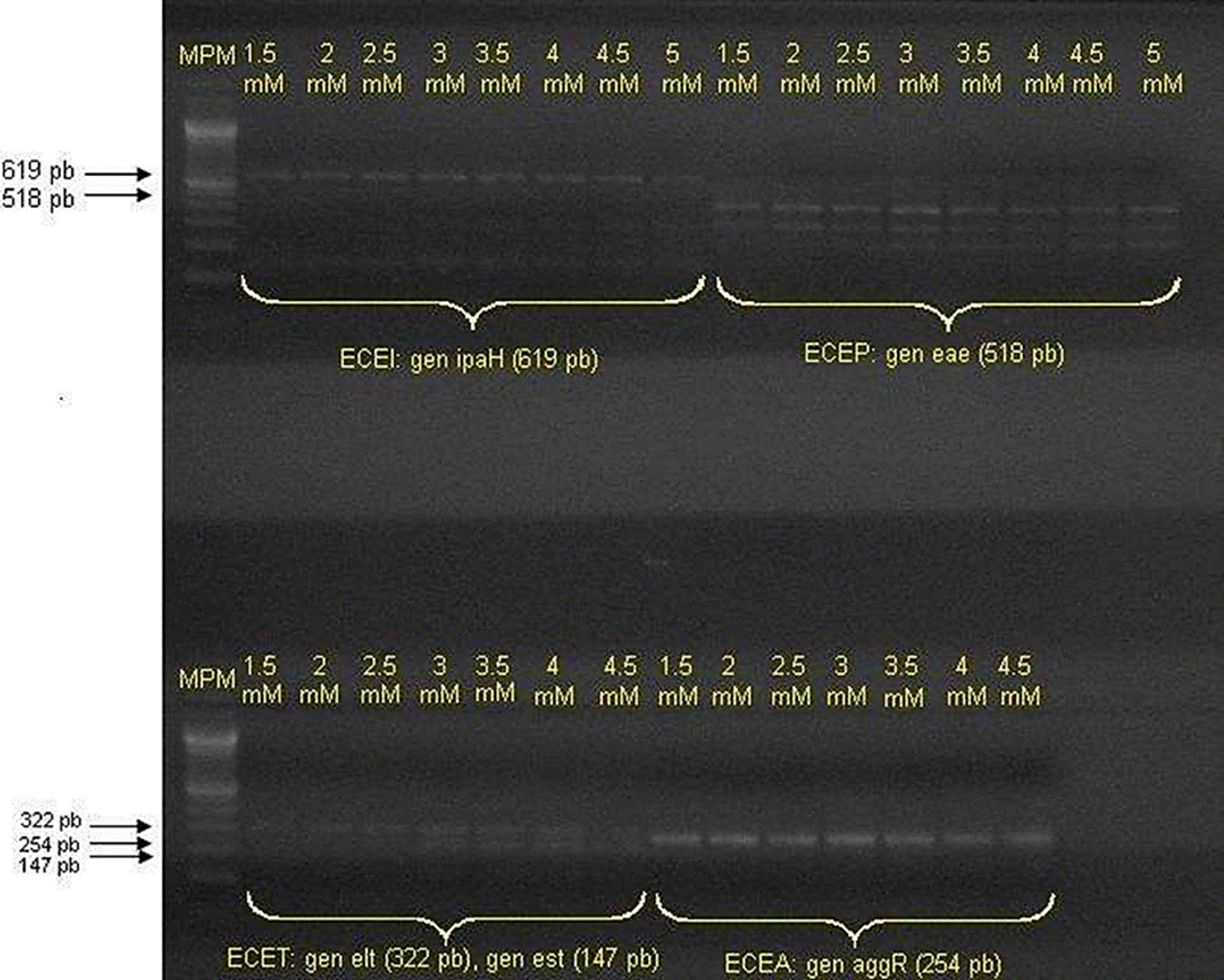
Curve of concentrations MgCl2 for PCR optimization.
Figure 1 Amplifications of Multiplex PCR selected reference strains.
Table 3 Standardized conditions and components for PCR reactions.
| Protocol stage and conditions | McDaniels et al. (1996) | Toma et al. (2003) | Fratamico et al. (2000) |
|---|---|---|---|
| Preheating | 94 °C 1 min | 94 °C 2 min | 94 °C 1 min |
| Desnaturalización | 94 °C 20 s | 94 °C 20 s | 58 °C 1 s |
| Alignment | 55 °C 1 min | 58 °C 1 min | 55 °C 1 min |
| Extension | 72 °C 1 min | 72 °C 1 min | 72 °C 1 min |
| Final extension | 72 °C 10 min | 72 °C 10 min | 72 °C 7 min |
| Components of the reaction mixture |
Polymerase buffer 1.25X MgCl2 2.5 mM dNTPs mix 0.2 mM Of each primer Forward and Reverse gadA/B 0.5 mM Taq Polymerase 2.5 U |
Polymerase buffer 1 X MgCl2 1.5 mM dNTPs mix 0.2 mM Of each primer Forward and Reverse 0.5 mM AL 65 0.25 mM SK1 0.25 mM ipaIII 0.25 mM LTL 0.25 mM est 0.25 mM ipaH 0.25 mM aggR Taq Polymerase 2.5 U |
Polymerase buffer 1X MgCl2 3.0 mM dNTPs mix 0.4 mM Of each primer Forward and Reverse 0.5 mM de stx1, stx2 y hly933 except 0.25 mM by eaeA Taq Polymerase 2.5 U |
| 50 ng of DNA contained in: | 5 µL | 5 µL | 5 µL |
| Reaction volume: | 50 µL | 50 µL | 50 µL |
* Reactions were carried out in a Thermal Cycler (Techne®, Bibby Scientific Ltd, U.K.).
Suspect strains for E. coli obtained for both methods of the protocol proposed by the FDA (2011) were submitted to the PCR proposed by McDaniels et al., (1996), for the detection of gadA/B gene, which encodes for glutamate decarboxylase, which is used for the confirmation of E. coli (Figure 2). The gadA/B strains that resulted to be positive were identified by the protocol proposed by Toma et al. (2003), to identify genetic determinants attributed to EIEC, EPEC, ETEC, EHEC and EAEC. The procedure consists in the detection of PCR products specific for each pathotype, by means of a multiplex PCR. In the same way, the strains confirmed by the protocol of MacDaniels et al. (1996) were submitted to multiplex PCR for EHEC proposed by Fratamico et al. (2000); which allowed amplifying four virulence factors for this pathotype (Figure 3). The oligonucleotides used were shown on Table 4.
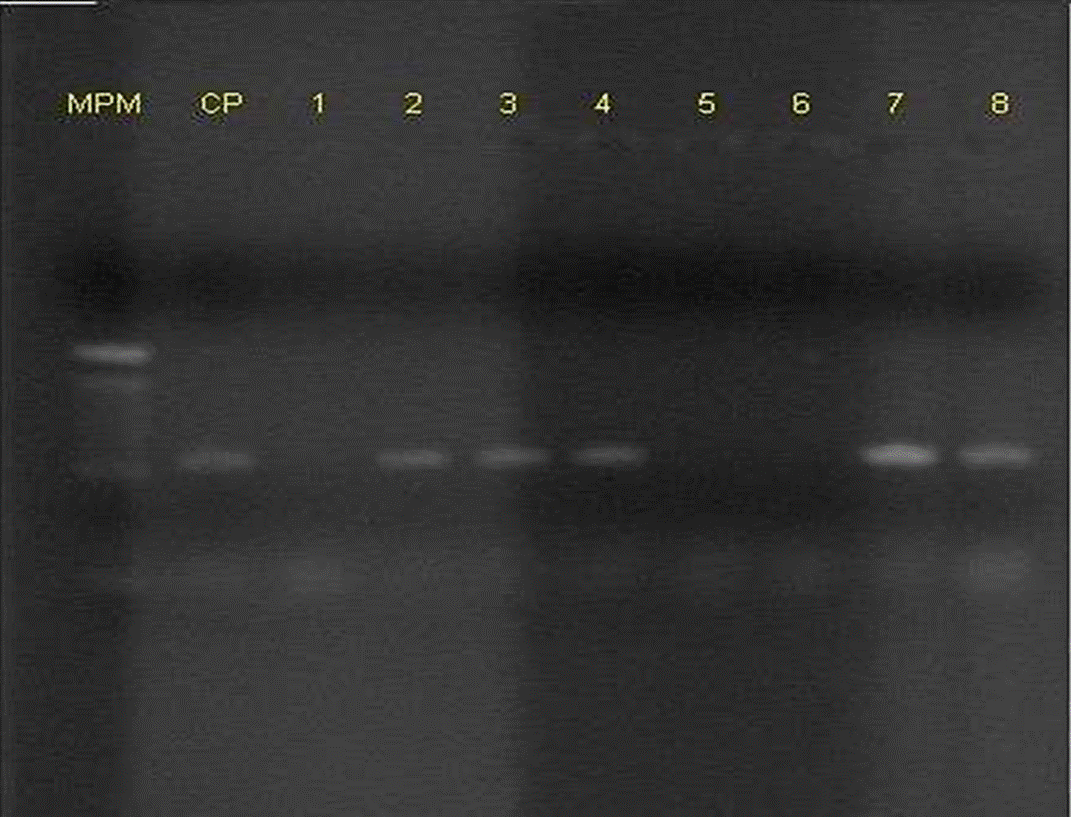
Figure 2 Representative gel showing multiplex PCR amplification of gadA/B in Escherichia coli enterotoxigenic (Positive Control, CP) and some strains (line 1 to 8).
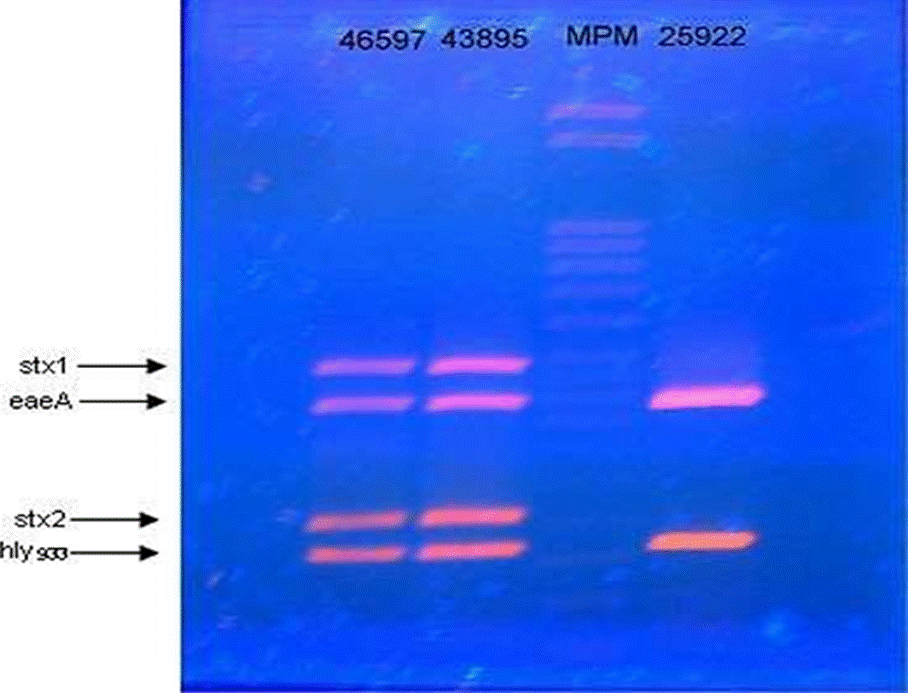
Figure 3 Gel showing multiplex PCR amplification of Escherichia coli enterohemorrhagic ATCC 46597, Escherichia coli enterohemorrhagic ATCC 43895 and, Escherichia coli enteropathogenic ATCC 25922.
Table 4 Oligonucleotides used in the protocols for PCR.
| Oligonu-cleotides | Sequence ( 5’ to 3’) | Target gene | Product Size (bp) | Encoding protein | Genus and species or patotype that it detects | Source |
|---|---|---|---|---|---|---|
| gadAB F | ACCTGCGTTGCGTAAATA | gadA/B | 670 | Glutamate decarboxylase | E. coli | McDaniels et al. (1996) |
| gadAB R | GGGCGGGAGAAGTTGATG | |||||
| AL65 | TTAATAGCACCCGGTACAAGCAGG | est | 147 | Thermostable enterotoxin | ETEC | Hornes et al. (1991) |
| AL125 | CCTGACTCTTCAAAAGAGAAAATTAC | |||||
| LTL | TCTCTATGTGCATACGGAGC | elt | 322 | Thermostable enterotoxin | ETEC | Tamanai - Shacoori et al. (1994) |
| LTR | CCATACTGATTGCCGCAAT | |||||
| ipaIII | GTTCCTTGACCGCCTTTCCGATACCGTC | ipaH | 619 | H antigen invasion plasmid | EIEC | Sethabutr et al. (1993) |
| ipaIV | GCCGGTCAGCCACCCTCTGAGAGTAC | |||||
| aggRksl | GTATACACAAAAGAAGGAAGC | AggR | 254 | Transcriptional activator for AAT / I expression | EAEC | Ratchtrachenchai et al. (1997) |
| aggRkas2 | ACAGAATCGTCAGCATCAGC | |||||
| SLTI-F | TGTAACTTGAAAGGTGGAGTATACA | stxl | 210 | Shiga toxin Type 1 | EHEC | Meng et al. (1997) |
| SLTI-R | GCTATTCTGAGTCAACGAAAAATAAC | |||||
| SLTII-F | GTTTTTCTTCGGTATCCTATTCC | stx2 | 484 | Shiga toxin Type 2 | EHEC | Meng et al. (1997) |
| SLTII-R | GATGCATCTCTGGTCATTGTATTAC | |||||
| AE22 | ATTACCATCCACACAGACGGT | eaeA | 397 | Intimate | E P E C , EHEC | Fratamico et al. (1995) |
| AE20-2 | ACAGCGTGGTTGGATCAACCT | |||||
| MFSI-F | ACGATGTGGTTTATTCTGGA | hly933 | 166 | Hemolysin (hlyA) | E P E C, EHEC | Fratamico & Strobaugh (1998) |
| MFSI-R | CTTCACGTCACCATACATAT |
The five pathotypes were analyzed by comparing the sizes of the PCR products of the samples with those of the control strains.
Results and Discussion
The 76 isolations were obtained from 15 of the 100 samples analyzed, from which 25 were isolated by means of the method for EHEC proposed by the FDA (2011) and 51 strains by the method for the other pathotypes.
A total of 55 strains (72.36 %) of the 76 isolated, amplified for the gadA/B gen, which were submitted to a multiplex PCR for detecting virulence factors for EHEC and the rest of E. coli pathotypes. It was found that 7.27 % (4/55) of the strains amplified the aggR gen of 254 pb, which encodes for the transcriptional activator of the expression of AAT/I of EAEC, and that 41.81 % (23/55) amplified for the elt gen of 322 pb that encodes for thermolabile enterotoxin of ETEC. No PCR products were identified for EPEC AND EIEC. On the other hand, of the 55 strains of E. coli confirmed and approved by multiplex PCR for EHEC, none of them showed PCR products that encode for virulence factors for this pathotype.
In relation with the amplification of virulence factors for EHEC, the control strain used amplified for type 1 Shiga toxin and type 2 Shiga toxin (stx2). It is important to highlight that this protocol had already been approved by Varela-Hernández et al. (2007), finding that the optimal concentration for the amplification of these virulence factors is 3.0 mM of MgCl2.
Once a PCR analysis was realized for the different protocols of PCR for pathogenic E. coli, it was revised from which samples came the positive strains.
The results of frequency and virulence factors amplified for this pathotype were shown in Table 5.
Table 5 Frequency of samples from which Escherichia coli pathotypes and amplified genes were isolated.
| Pathotype | Amount of positive samples | Number of positive strains/total strais (percentage) | Amplified virulence factor |
|---|---|---|---|
| EAEC | 2 | 4/55 (7.27 %) | aggR |
| ETEC | 13 | 23/55 (41.18 %) | elt |
| EIEC | 0 | 0/55 | - |
| EPEC | 0 | 0/55 | - |
| EHEC | 0 | 0/55 | - |
After the association of the results of the identification of pathotypes in strains with respect to sampled months and samples analysis, the results were shown on Table 1. It is important to mention that at national level, there are only a few studies of this type. Fifteen percent (15/100) of the samples resulted to be positive for some E. coli pathotype, all of them were detected in the warm months in the city of Guadalajara, Jalisco, in agreement with Varela-Hernández et al. (2007) and Cagney et al. (2004), who reported a seasonal behavior, in researches on prevalence in minimally processed fresh beef, where they found that the probability to isolate EHEC was greater in spring-summer than in fall-winter.
Villaseca et al. (2005) described some outbreaks associated with pathogenic E. coli registered in hospitals from Central America countries, however, no reference was made to the infection vehicle of the patients. A higher frequency of patients infected by the pathogen was declared, who attend hospitals during the warm months (April to June), data coinciding with the frequency of E. coli pathotypes, according to the sampling month, found in this study.
Particularly in this zone of western Mexico, it coincided that in May, the highest annual average temperature was observed to be 33 ± 2.1 ºC, with the highest frequency of isolation of E. coli pathotypes (CLIMATE-DATA, 2019). This data is important to be considered since in the butcher’s shops of the country, meat is exposed to the public at room temperature.
Barlow et al. (2006) collected 285 samples of ground beef in 10 occasions of 31 butcher’s shops for one year in Australia; of which 16 % (46/285) were presumptive for EHEC by means of the method proposed by the FDA (2011), similar frequency to that found in the present study (15 %); although only one sample per shop was collected in this study. Barlow et al. (2006) isolated 63 STEC strains of which 5 % (3/63) encode for stx1, 55 % (35/63) for stx2 and 65 % (41/63) for intimine, contrary to the present study in which of a total of 25 presumptive strains of EHEC, none of them amplified for any virulence factor, specifically for the approved ones.
On the other hand, Etcheverría et al. (2010) detected samples positive of STEC, between 12.34 % and 18.64 % in isolated samples of carcasses in sanitary control cabins of slaughterhouses in Argentina and 24.5 % in meat coming from butcher’s shops; which contrasts with 15 % of the two pathotypes found in the present study. As well, Cagney et al. (2004), in the Republic of Ireland, in a study with similar characteristics to ours, found that from 140 samples of ground beef obtained from shops, none detected EHEC, which coincides with our results. In this context, it is necessary to implement hygiene measures and health programs that guarantee the innocuousness of the meat starting from its acquisition, transport and storing in the shop, that avoid or minimize crossed contamination and the growth of these microorganisms, not exempting the thermic elimination of the pathogen when cooking the meat.











 texto en
texto en 



