Introduction
Heteromys irroratus, the Mexican spiny pocket mouse, is a species widely distributed in México and in a small portion of the United States. In México, it is found on the Mexican Plateau and in adjacent areas, and along the main mountain systems and coastal slopes (Genoways 1973; Dowler and Genoways 1978). Altitudinally, H. irroratus occurs from sea level, across the coasts of Tamaulipas and Veracruz to nearly 3,050 m in the highlands of the Sierra Madre de Sur, in México. It can be found mainly in xerophilous scrubland thorny forest, although it also occurs in coniferous and oak forests, and agricultural, and grazing lands (Dowler and Genoways 1978). Currently, seven subspecies are recognized (H. irroratus alleni, H. i. bulleri, H. i. guerrerensis, H. i. irroratus, H. i. jaliscensis, H. i. texensis, and H. i. torridus; Genoways 1973; Dowler and Genoways 1978). Of the seven, H. i. bulleri has the most restricted distribution.
Heteromys i. bulleri was described by Thomas (1893) from La Laguna, Sierra de Juanacatlán, Jalisco (México) and is only known from seven specimens collected in the mid-1960’s; two from the type locality and five more from the vicinity of Soyatlán del Oro, Jalisco (Genoways 1973). These two localities are approximately 60 km from each other in the western portion of the state (Figure 1). Although this taxon was described almost 130 years ago, little is known about its natural history.
Recent surveys in the Sierra de Manantlán Biosphere Reserve, in the southwestern portion of Jalisco (México), uncovered the existence of a new population of H. irroratus. Although H. i. jaliscensis has been reported from localities not far from the Biosphere Reserve and its type locality is about 50 km distant from Sierra de Manantlán, specimens representing this population did not resemble H. i. jaliscensis morphologically. External and cranial measurements of individuals of the new population (Table 1) are larger than the ones reported for individuals of H. i. jaliscensis (including samples from its type locality; Genoways 1973). Therefore, considering that H. i. bulleri also occurs in Jalisco, although in a different area, our aim was to assess the subspecific assignment of the Manantlán Biosphere Reserve specimens. Due to the fact that there are only two specimens of H. i. bulleri from the type locality (holotype at the British Museum (Natural History), London, United Kingdom, and a topotype at the National Museum of Natural History, United States) and a direct comparison with them was not possible, field work was conducted at the type locality of H. i. bulleri to obtain samples to compare with specimens of the new population using Cytochrome b (Cyt b) sequences. In this study, we report the findings of the molecular identification of the novel samples and their distinctiveness, both in terms of genetic divergence and phylogenetic placement, compared to available sequence data for H. irroratus.
Material and methods
Eight specimens of the new population of Heteromys irroratus were collected at Las Joyas Scientific Station, located in the Sierra de Manantlán Biosphere Reserve, Municipality of Autlán, Jalisco, México (19° 35.443’ N, -104° 16.913’ W) at 1,957 masl. The area is characterized by pine-oak forest and cloud forest, but spiny pocket mice were captured near a field of Zea diploperennis, an endemic perennial species of teosinte or wild corn. Voucher specimens (preserved in alcohol) were deposited in the Colección Zoológica de Vertebrados, Centro Universitario de la Costa Sur, Universidad de Guadalajara (CVUDG: 1675, 1676, 1710, 1711, 1713, 1714, 1715, 1716). An additional individual of H. irroratus was collected at La Laguna, Sierra de Juanacatlán, Municipality of Mascota, Jalisco, México (20° 37.687’ N, -104° 43.752’ W), at 2,050 masl, which represents the type locality of the subspecies H. i. bulleri. This specimen was caught in secondary vegetation within a pine-oak forest next to a lake. The voucher specimen (preserved as skin and squeleton) is stored in the Colección de Mamíferos del Centro de Investigación en Biodiversidad y Conservación de la Universidad Autónoma del Estado de Morelos (CMC 3590). Sequences generated as part of this study were uploaded to GenBank and accession numbers are included in Appendix 1. Capture and handling methods followed the animal care and use guidelines of the American Society of Mammalogists (Sikes et al. 2016).

Figure 1 Map illustrating collecting records of Heteromys irroratus bulleri (dots) and H. i. jaliscensis (squares). 1) La Laguna, Sierra de Juanacatlán (H. i. bulleri type locality). 2) Soyatlán del Oro (Genoways 1973). 3) Sierra de Manantlán Biosphere Reserve (current study). 4) Vicinity of Ameca (molecular data from specimens collected at this locality are included in current study as H. i. jaliscensis). 5) Las Canoas (H. i. jaliscensis type locality).
Three external and nine cranial measurements, described by Genoways (1973), were recorded for seven adult specimens of H. irroratus from Sierra de Manantlán (four females and two males) and one adult specimen from Sierra de Juanacatlán (one female; see Table 1). External measurements were obtained from collecting field catalogs, while cranial measurements were recorded by means of a digital caliper with accuracy of 0.1 mm. Discrimination of age categories that represent adult individuals was based on toothrow wear patterns described by Genoways (1973). Measurements were compared to data recorded by Genoways (1973) for individuals of H. i. bulleri and H. i. jaliscensis occuring near Sierra de Manantlán and Sierra de Juanacatlán. Because Genoways (1973) documented that female specimens of H. i. jaliscensis are smaller than males, comparison of measurements among specimens of H. irroratus from Sierra de Manantlán and Sierra de Juanacatlán and H. i. jaliscensis were performed by sex. Box plots of external and cranial measurements were generated using STATISTICA v.8.0 (StatSoft 2007).
Total genomic DNA from the newly collected specimens was extracted from liver tissue preserved in 95 % ethanol following the procedure described by Fetzner (1999). Four microliters of DNA were electrophoresed on 1.75 to 2.0 % agarose gels stained with SYBR Green to visualize quality of genomic DNA. MVZ05 and MVZ14 primers (Smith and Patton 1993; Arellano et al. 2005) were used to amplify the Cyt b gene (1140 bp). PCR master mix contained: 1.0 μl of template DNA (approximate concentration estimated on a 2 % agarose gel), 1 μl of deoxynucleosidetriphosphates (10 mM), 5 μl of 10x Taq buffer containing MgCl2, 1 μl of each primer (100 mM concentration), 40.7 μl of distilled water, and 0.3 μl of Taq polymerase (5 U/μl; Promega Corp., Madison, Wisconsin) for a 50 μl final volume. Standard amplification conditions consisted of 2 to 4 min at 94 °C for initial denaturation (1 cycle); then, 1 min at 94 °C for denaturation, 1 min at 45 °C for annealing, and 1 min at 72 °C for extension (35 cycles); lastly, 5 min at 72 °C for final extension (1 cycle). Four microliters of PCR-amplified product were assayed by electrophoresis on a 2 % agarose gel. PCR products were purified with a Gene-Clean PCR purification kit (Bio 101, La Jolla, California). Sequencing reactions of purified PCR products were done with the Perkin-Elmer ABI PRISM dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, California). Excess dye terminator was removed using a Sephadex 50G solution (3 g/50 ml H2O). Light- and heavy-strand sequences were determined with an ABI 3100 automated sequencer (Applied Biosystems) housed in the DNA Sequencing Center at Brigham Young University. Final sequences were edited using BioEdit v.7.0.8.0 (Hall 1999). GenBank sequences accession numbers are MT709150, 709151, 709152, 709153, 709154, 709155, 709156, T709157, and (see Appendix 1 for correspondence of accession numbers and voucher collecting information).
New sequences were added to a matrix together with 33 Cyt b sequences of H. irroratus available from GenBank and representing the following subspecies: H. i. alleni, H. i. guerrerensis, H. i. irroratus, H. i. jaliscensis, H. i. torridus, and H. i. texensis (first described by Rogers and Vance 2005; Appendix 1). Also, sequences of H. pictus accessible from GenBank were downloaded and used as the outgroup in phylogenetic analyses (Appendix 1). Our final matrix included 786 bp of Cyt b.
Sequence alignment was done with BioEdit v.7.0.8.0 (Hall 1999). Hypotheses concerning phylogenetic relationships among sequences were estimated using Maximum Likelihood (ML) and Bayesian Inference (BI) employing IQ-TREE v.1.6.10 (Kalyaanamoorthy et al. 2017; Nguyen et al. 2015) and MrBayes v.3.2.2 (Ronquist et al. 2012) on XSEDE, respectively. The model of DNA evolution most appropriate for our data was selected using jModelTest2 on XSEDE (Darriba et al. 2012) under the Bayesian information criterion (BIC). To assess if data needed to be partitioned by codon position, we employed ModelFinder (Kalyaanamoorthy et al. 2017). The transition model 2 with invariable sites and rate heterogeneity (TIM2+I+G4; Tamura and Nei 1993) was determined as the best-fit model of nucleotide substitution (πA = 0.288, πC = 0.248, πG = 0.142, and πT = 0.323; rAC/AT = 1.000/1.000, rCG/GT = 1.000/1.000; I = 0.475; α = 0.224), and the data were not partitioned. For the ML analysis, tree searches were performed employing IQ-TREE default search parameters. Branch support was assessed through ultrafast bootstrap (UB) with 1,000 replicates (Hoang et al. 2018). For the BI analyses, runs were specified with four of MCMC chains and 50,000,000 generations, with a sample frequency of 1,000 generations. This resulted in 50,000 samples from the posterior probability (PP) distribution. Burn-in value was set to 10,000. This resulted in 50,000 samples and the burn-in value was set to 10,000. The posterior probability (pP) was computed for individual branches by constructing a majority-rule consensus with the trees not discarded as burn-in. Once the main phylogenetic haplogroups were identified, uncorrected (p-distances) and Kimura-2-parameters (K2P; Kimura 1980) genetic distances were calculated employing Mega v.7.0.3 (Kumar et al. 2016). The former genetic measurement is provided for further comparison to other mammal taxa. The latter genetic distance was compared between selected sequences of H. irroratus since it has been widely used to compare levels of DNA sequence divergence in mammals under the Genetic Species Concept framework (Bradley and Baker 2001; Baker and Bradley 2006).
Results
External and cranial measurements among specimens of the new population of H. irroratus from Sierra de Manantlán and individuals of H. i. jaliscensis showed overlap in the ranges displayed by most variables for both females and males (Table 1; Figures 2, 3). On average, the four adult females of H. irroratus from Sierra de Manantlán are larger than the females of H. i. jaliscensis. This is particularly evident for three external measurements (total length, length of tail, length of hind foot; Figures 2a-c) and five cranial variables (greatest length of skull, mastoid breadth, length of nasals, length of maxillary toothrow, depth of braincase; Table 1; Figures 2d-j). The two adult males from Sierra de Manantlán showed larger mean values for three cranial measurements (mastoid breadth, length of nasals, and depth of braincase) compared to males of H. i. jaliscensis (Table 1; Figures 3f-j). The only adult female specimen of H. irroratus from Sierra de Juanacatlán, also displayed larger values for all three external and seven cranial measurements compared to average values depicted by females of H. i. jaliscensis (Table 1; Figures 2a-j). Mean lengths of the adult females of H. irroratus from Sierra de Manantlán and Sierra de Juanacatlán were more similar to the mean measurements of specimens of H. i. bulleri reported by Genoways (1973), particularly two external variables (total length and length of tail; Table 1; Figures 2a, 2b) and one cranial measurement (length of nasals; Table 1; Figure 2g).
Table 1.External and cranial measurements (in mm) for specimens of H. i. jaliscensis and H. i. bulleri. When more than one individual was available, the range is shown for each variable. Mean values are in parentheses. Sierra de Manantlán (SM), Sierra de Jaunacatlán (SJ).
| Genoways (1973) | This study | |||||
|---|---|---|---|---|---|---|
| External and cranial measurements | H. i. jaliscensis | H. i. bulleri* | H. i. bulleri SM | H. i. bulleri SJ | ||
| Females | Males | Females | Females | Males | Female | |
| (n = 51) | (n = 38) | (n = 3) | (n = 4) | (n = 2) | ( n = 1) | |
| Total length | 207.0-251.0 (226.9) | 212.0-272.0 (238.3) | 245.0-250.0 (247.4)** | 226.0-255.0 (245.5) | 228.0-249.0 (238.0) | 249 |
| Length of tail | 101.0-131.0 (114.1) | 103.0-140.0 (118.7) | 127.0-128.0 (127.4)** | 113.0-134.0 (125.0) | 125 | 128 |
| Length of hind foot | 25.0-30.0 (27.3) | 26.5-32.0 (28.9) | 27.0-32.0 (29.3)** | 26.0-32.0 (29.2) | 25.9-30.0 (27.8) | 33 |
| Greatest length of skull | 30.1-33.2 (31.6) | 30.4-35.1 (32.6) | 33.5-34.1 (33.8) | 31.8-33.5 (32.7) | 33.7 | 32 |
| Interorbital constriction | 7.2-8.6 (7.8) | 7.2-9.0 (8.0) | 8.2-8.7 (8.5) | 7.6-8.1 (8.0) | 7.7-7.9 (7.8) | 8.7 |
| Mastoid breadth | 13.8-14.7 (14.2) | 13.8-15.9 (14.8) | 14.8-15.5 (15.1) | 14.4-15.4 (14.8) | 14.8-15.3 (15.0) | 14.6 |
| Length of nasals | 11.0-13.5 (12.1) | 11.4-13.4 (12.3) | 13.2-14.1 (13.6) | 13.2-14.6 (14.1) | 14.4-14.6 (14.5) | 13.9 |
| Length of rostrum | 12.9-14.9 (13.8) | 12.2-15.7 (13.7) | 15.3** (15.3) | 12.8-14.4 (13.9) | 14.1-14.5 (14.3) | 13.6 |
| Length of maxillary toothrow | 4.4-5.4 (4.8) | 4.8-5.8 (5.3) | 5.9-6.0 (5.9) | 5.5-5.9 (5.6) | 5.2-5.3 (5.2) | 5.3 |
| Depth of braincase | 8.2-9.1( 8.6) | 8.2-9.3 (8.7) | 9.4-10.0 (9.7)** | 9.2-9.8 (9.5) | 9.4-9.7 (9.5) | 9.4 |
| Interparietal width | 7.3-9.5 (8.3) | 7.9-9.8 (8.7) | 5.4-8.0 (6.7) | 7.4-7.8 (7.8) | 7.4 | 8.6 |
| Interparietal length | 2.5-4.4 (3.2) | 3.0-4.4 (3.6) | 3.2-3.9 (3.6) | 3.7-4.2 (3.9) | 3.9 | 4.1 |
*Samples included two adult females from La Laguna, Sierra de Juanacatlán (type locality) and one adult female from Soyatlán del Oro.
**Data preceded by two asterisks were recorded for only two specimens.
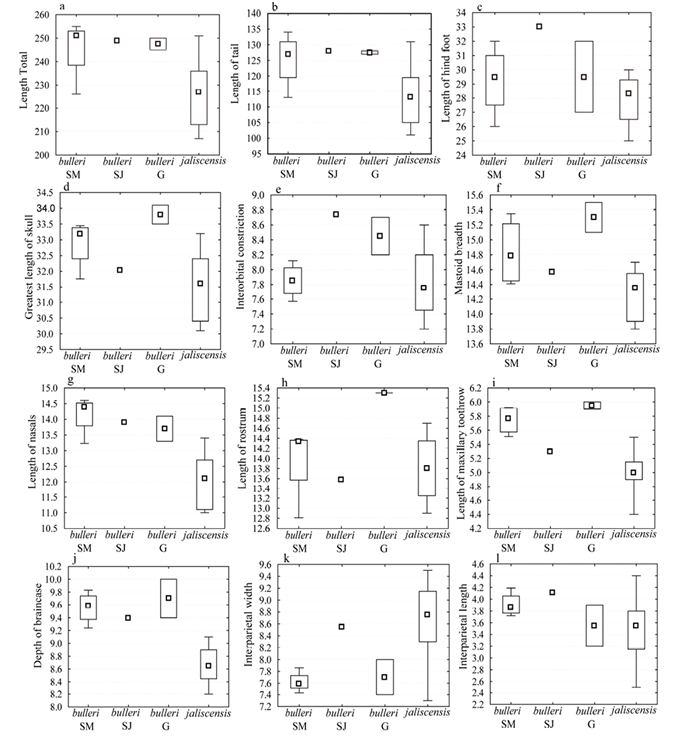
Figure 2 Box plots of female measurements of individuals of H. i. bulleri from Sierra de Manantlán (bulleri SM), H. i. bulleri from Sierra de Juanacatlán (bulleri SJ), H. i. bulleri from Genoways (1973; bulleri G), and H. i. jaliscensis from Genoways (1973; jaliscensis). Median (small squares), 25% to 75% confidence interval (rectangles), and minimum and maximum values (whiskers).
Phylogenies derived from ML (Figure 4) and BI (not shown) were highly congruent. Sequences from Sierra de Manantlán grouped together with high nodal support (UB = 91 and PP = 0.99). In turn, the sequence from Sierra de Juanacatlán (CMC 3590) joined those sequences from Sierra de Manantlán forming a strongly supported haplogroup (UB = 100 and PP = 1.0; Figure 4). The H. i. bulleri haplogroup was positioned as a divergent sister group to the other sequences of H. irroratus. The remaining sequences of H. irroratus formed a monophyletic group with strong nodal support values (84 / 0.99). This clade contained representatives of the other six subspecies of H. irroratus (alleni, guerrerensis, irroratus, jaliscensis, texensis, and torridus). Sequences of each subspecies formed separate haplogroups, with the exception of H. i. alleni. Sequences representing this subspecies split into two independent lineages, H. i. alleni (1) and H. i. alleni (2). Although two divergent lineages were also found within H. i. texensis, they are sister to each other. In summary, we identified a total of eight haplogroups within H. irroratus.

Figure 3 Box plots of male measurements of individuals of H. i. bulleri from Sierra de Manantlán (bulleri SM) and H. i. jaliscensis from Genoways (1973; jaliscensis). Median (small squares), 25% to 75% confidence interval (rectangles), and minimum and maximum values (whiskers).
Pairwise K2P genetic distances (Kimura 1980) between localities 1 and 3 of H. i. bulleri (Figure 1) ranged between 0.63 % and 1.02 % with a mean of 0.84 % (Table 2). In contrast, the mean K2P genetic distance between individuals of H. i. bulleri and the other haplogroups ranged from 10.05 % (H. i. jaliscensis) to 11.94 % (H. i. irroratus). Among all haplogroups of H. irroratus, except H. i. bulleri, K2P distances ranged between 1.59 % (H. i. guerrerensis-H. i. irroratus) and 8.34 % (H. i. jaliscensis-H. i. alleni (2); Table 2).
Table 2 Pairwise Kimura-2-parameter distances (below the diagonal) and p-distances (above the diagonal), between samples of Heteromys irroratus based on 786 bp of the Cytochrome b gene.
| Taxon | H. i. irroratus | H. i. texensis | H. i. alleni (1) | H. i. jaliscensis | H. i. alleni (2) | H. i. guerrerensis | H. i. torridus | H. i. bulleri |
|---|---|---|---|---|---|---|---|---|
| H. i. irroratus | 0.0694 | 0.0635 | 0.0705 | 0.0734 | 0.0157 | 0.0564 | 0.1079 | |
| H. i. texensis | 0.0742 | 0.0407 | 0.0505 | 0.0744 | 0.0637 | 0.0405 | 0.1016 | |
| H. i. alleni (1) | 0.0675 | 0.0423 | 0.0364 | 0.0657 | 0.0619 | 0.0233 | 0.0930 | |
| H. i. jaliscensis | 0.0751 | 0.0527 | 0.0375 | 0.0776 | 0.0636 | 0.0449 | 0.0927 | |
| H. i. alleni (2) | 0.0789 | 0.0800 | 0.0701 | 0.0834 | 0.0725 | 0.0623 | 0.1049 | |
| H. i. guerrerensis | 0.0159 | 0.0678 | 0.0659 | 0.0674 | 0.0780 | 0.0534 | 0.1025 | |
| H. i. torridus | 0.0595 | 0.0421 | 0.0238 | 0.0467 | 0.0662 | 0.0563 | 0.0973 | |
| H. i. bulleri | 0.1194 | 0.1116 | 0.1013 | 0.1005 | 0.1160 | 0.1129 | 0.1064 |
Discussion
This study focuses on the molecular taxonomic identification of individuals of a new populations of H. irroratus from Sierra de Manantlán Biosphere Reserve, a locality in southern Jalisco (México), and its phylogenetic placement within this group. Our initial identification assessment involved the comparison of external and cranial measurements to H. i. jaliscensis and H. i. bulleri, two subspecies of this taxon occurring not far from this new collecting locality. Yet, ranges of measurements between adult females from Sierra de Manantlán and those of H. i. jaliscensis overlap, the former are on average larger than the latter (Table 1, Figures 2, 3). Although differences in length between males from Sierra de Manantlán and H. i. jaliscensis were not evident for most variables, specimens from Sierra de Manantlán displayed larger mastoid breadth, length of nasals, length of rostrum, and depth of braincase. On the other hand, measurements of females from Sierra de Manantlán were similar to those of H. i. bulleri reported by Genoways (1973). These results, principally the data recorded for females, preliminary suggest that specimens of the new population of H. irroratus from Sierra de Manantlán were not representatives of H. i. jaliscensis, but rather, represented the subspecies H. i. bulleri. In addition, the female specimen collected at the type locality of H. i. bulleri (La Laguna, Sierra de Juanacatlán), exhibited measurements that better agree with those reported for H. i. bulleri than for H. i. jaliscensis (Genoways, 1973 Genoways 1973), suggesting that this individual also belongs to the former taxon. This is in agreement with the findings reported by Genoways (1973) who concluded that H. i. bulleri can be discriminated from H. i. jaliscensis (and other subspecies) by its larger external and cranial size.

Figure 4 Maximum likelihood phylogenetic tree of Heteromys irroratus sequences based on 786 bp of the Cytochrome b gene (lnL = -3357.567). Posterior probability estimates are shown above branches and bootstrap values are below branches.
Although, it was not possible to make a comparison with type specimens of H. i. bulleri, color patterns (fresh pelage) of the individual collected at the type locality (upperparts gray mixed with black and ochraceous hairs; sides with a pale ochraceous line; underparts white; feet white; ears dusky with a white edge; tail bicolor, brown above and white below) match those described for H. i. bulleri (Thomas 1893; Goldman 1911), supporting the assignment of this specimen to this taxon. Although Heteromys pictus could also occur at the type locality of H. i. bulleri, it can be distinguished from H. irroratus by the presence of six plantar tubercles on each hind foot (compared to five plantar tubercles displayed by H. irroratus; Genoways 1973) and overall smaller size. The fact that all the specimens of Heteromys from Sierra de Manantlán and Sierra de Juanacatlán collected for our study possess five plantar tubercles on each hind foot, supports the hypothesis they are representatives of H. irroratus.
When comparing Cyt b sequence data between the sample from Sierra de Juanacatlán (type locality of H. i. bulleri) to any of the individuals from Sierra de Manantlán, the genetic distance values were < 1.02 % (mean 0.84 %), lower than the upper bound (1.8 %) of the intrasubspecific range reported by Bradley and Baker (2001) for Cyt b in rodents. This suggests a close affinity between individuals from the Sierras de Juanacatlán and Manantlán and supports that they represent the same subspecies. This hypothesis is also supported by the fact that all these sequences formed a single haplogroup with strong nodal support. Since the sample from Sierra de Juanacatlán represents the type locality of H. i. bulleri, we propose that specimens from the Sierra de Manantlán also represent H. i. bulleri. Certainly, confirmation of this findings would be desirable by analyzing additional data such as nuclear markers and morphological features.
Although we did not have molecular data for H. i. jaliscensis from its type locality (Las Canoas, Jalisco) to confirm that specimens from Sierra de Manantlán do not belong to that subspecies, we included representatives of H. i. jaliscensis from Ameca, Jalisco. Samples of H. irroratus from these two localities (Las Canoas and Ameca), were analyzed morphologically by Genoways (1973) and regarded as the same subspecies. The levels of genetic differentiation between H. i. jaliscensis and specimens from Sierra de Manantlán were comparatively high (10.05 %; Table 2), supporting the hypothesis that they represent distinct subspecies.
Our phylogenetic hypothesis places H. i. bulleri as the sister group to all other forms of H. irroratus (Figure 4). The reciprocal monophyly of these two clades was well supported (H. i. bulleri = 100/1.0; other H. irroratus = 84/0.98). Likewise, the high degree of genetic divergence among sequences of these two clades was documented with K2P; in any pairwise comparison between these two groups, genetic distances exceeded 10 %. This level of Cyt b genetic differentiation can be indicative of distinct species (Bradley and Baker 2001; Baker and Bradley 2006).
Although H. i. bulleri is currently recognized as a subspecies (Genoways 1973), it was originally described as a species by Thomas (1893:330) because it had a “skull strong and stoutly built, differing mainly from that of H. alleni in its much greater size, and from that of H. irroratus by its differently shaped interparietal”. The specific status of H. i. bulleri was supported by Goldman (1911:62) who wrote: “In external appearance it resembles alleni and jaliscensis, but the cranial characters, especially the decidedly smaller size and more triangular shape of the interparietal, distinguish it from either”. Genoways (1973), based on a morphological analysis of all known forms of the Mexican spiny pocket mouse, found that samples of H. i. bulleri possessed deeper braincases, compared to other groups of H. irroratus. Heteromys i. bulleri also was separated from all other samples of H. irroratus in a Principal Component Analysis. In addition, a UPGMA phenogram showed that, at least for the sample from Soyatlán del Oro, is quite distinct from the rest of samples representing the Mexican spiny pocket mouse. Finally, Genoways (1973:106) observed that “the small size of the interparietal is rather unique” for bulleri, although he pointed out that this feature was shared with a population of H. i. alleni from Michoacán. However, he concluded that, due to the high variation of the shape and size of the interparietal bone in H. irroratus and other species in the genus, specific distinction of bulleri should not be based on that single character, and therefore, he relegated it to the subspecific level.
Although our molecular data represent a portion of a single mitochondrial gene, the level of genetic differentiation documented by this marker for H. i. bulleri relative to other samples of H. irroratus, suggest that it represents a distinct species-level taxon. However, we believe that additional data (both molecular and morphological) are necessary to further test the specific status of this taxon.
The fact that H. i. bulleri should be considered a distinct species supports previous views suggesting that H. irroratus is a species complex containing some species-level lineages (Rogers and Vance 2005). It has been demonstrated that individuals of H. irroratus from near Pátzcuaro, Michoacán, México (represented by our H. i. alleni (2) haplogroup; Figure 2) were genetically distinct and can be considered as a candidate species. Also, it was concluded that populations of H. irroratus from Guerrero and Oaxaca are genetically differentiated and may represent a second candidate species (Rogers and Vance 2005).
Since its description in 1893, only 16 specimens of H. i. bulleri have been collected: two reported at the time of its description, five more collected between 1956 and 1966, and nine more reported in this study were trapped between 2014 and 2020. The scarce number of voucher specimens collected in almost 130 years suggests that either the trapping efforts have been insufficient to properly sample it or this taxon is uncommon. The Sierra de Manantlán is a new locality for H. i. bulleri and represents a range extension of about 125 km southeast from the type locality and 80 km south from Soyatlán del Oro. Given that H. i. bulleri is distinctive molecularly and morphologically and coupled with its restricted distribution, it is crucial to pay attention to conservation issues surrounding this taxon.











 nueva página del texto (beta)
nueva página del texto (beta)


