Introduction
The Atoyac River originates in the Calpulalpan sierra in the state of Tlaxcala and along its course, through the state, it is fed by the Zahuapan River forming the Alto Atoyac sub-basin with an area of 2013 km2 (Carrera-Villacrés, 2007). The Atoyac River later finds its way into the state of Puebla and runs through its capital city before ending up on the Valsequillo dam. However, these rivers receive urban, industrial and agricultural effluents from the towns and industrial activities along with their course. The industrial activities are quite varied and they include the food, textile, chemical, petrochemical, pharmaceutical, metallurgic, electric, and automobile industries, among others (Sandoval-Villasana, Pulido-Flores, Monks, Gordillo-Martínez, & Villegas-Villareal, 2009). A previous study has assessed the quality of waters, sediments, and the genotoxic potential in the hydrological Atoyac-Zahuapan river system and the Avila Camacho dam (Mangas et al., 2005). The genotoxic potential of sediments and superficial water of this system has been confirmed with the plant Vicia faba (Villalobos-Pietrini, Flores, & Gómez, 1994; Juárez-Santacruz et al., 2012). Chromosomal aberrations and centromeric alterations were found in root tip cells of this plant after being exposed to the Atoyac River waters (ob. cit.) while the micronuclei frequency was increased after exposure to the river sediments in a recent study (Juárez-Santacruz et al., 2012). Moreover, metals such as lead and arsenic have been detected in the Alto Atoyac sub-basin and their concentrations were above the Mexican and international permissible limits (García-Nieto et al., 2011). Other studies have also revealed the number and location of effluents in the Atoyac River (Saldaña & Gómez, 2006; Sandoval-Villasana et al., 2009) and have established them as the main source of contamination. Furthermore, concentrations of Mn and Fe in waters of the Valsequillo dam were also above the permissible limits (Díaz, Bonilla, Tornero, Cabrera, & Corona, 2005). These waters are used for irrigation of nearby crops such as maize and alfalfa and their soils also have Pb concentrations above the norm (Larenas-Bazán, 2010). The most recent study on these river sediments and waters reported severe pollution that includes hydrocarbons, metals, POPs, endocrine disruptors and potentially carcinogenic compounds (Greenpeace, 2014). Therefore, this study aims to elucidate the microbial diversity in a river area where it has received all the major domestic and industrial effluents prior to the water storage in the Valsequillo dam. The identification of the microbial diversity will provide a clearer idea of their catabolic potential in those sediments.
Materials and methods
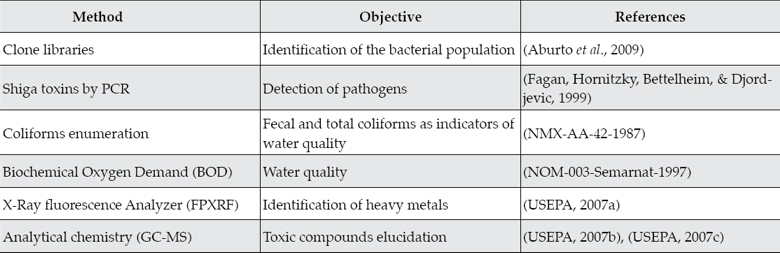
A list of the different methods used in this study is included in table 1.
Site description and sampling
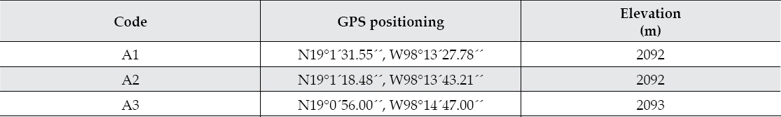
Sediment and water samples were collected from three different positions (A1-A3) located just prior the Valsequillo dam in central México. The river water at these positions has undergone wastewater treatment processes after receiving industrial and domestic effluents along the river course. The global positioning coordinates for the sampled sites and the relative position of the samples are indicated in table 2 and figure 1, respectively. The samples were collected during the dry season on 21 January 2013. Water samples were collected in sterile 1-litre glass bottles. Sediments from the bottom of the river were collected in 50 ml sterile centrifuge tubes. Then, they were immediately preserved in dry ice and protected from light.
Determination of elements in sediments
The sediments were dried to room temperature to prevent any volatilization. Each sample was homogenized and then a representative portion was milled with a mortar prior to passing it through a 60-mesh sieve before a further homogenization. The total element concentrations (As, Pb, Ca, Fe, K, Ti, Sr, Ba, Mn, Zr, Zn, Cu, Rb, Mo) were determined in 15 g of dried sediments following the Unites States Environmental Protection Agency Method 6200 (USEPA, 2007a), using a field portable X-Ray fluorescence Analyzer (FPXRF), NITON XL3t Thermo Scientific. Two reference materials were used to establish the method accuracy: the Standard Reference Material “NIST 2710” Montana Soil and the Reference Material unstratified glacial deposit Till-4. The accuracy percentage of all elements was in the 80 - 120 % range. Moreover, duplicate analyses were performed to establish the method precision and the relative difference percentage (% RDP) was always below 10%. To discard contamination interferences, blanks were analyzed between each interest sample. The analysis of clean quartz dioxide matrix (blank) was always below the detection limit for all elements.
Determination of organic compounds
The organic compounds were extracted from dried sediments by sonication Method 3550C, (USEPA, 2007b) using 3 g of soil and 95 ml of a mixture 1:1 n-hexane-ketone (Mallinckrodt, HPLC grade). The extracts were concentrated by roto-evaporation and changed to n-hexane as a solvent. The recovery percentage of this extraction method was validated using a soil sample with known concentrations of 17 reference compounds. The percentage of recovery of all compounds was higher than 90%. Regular controls with this soil were performed to ensure the quality of the data. The extracts were analyzed to identify organic compounds by gas chromatography Method 8270D (USEPA, 2007c) coupled with a mass spectrum detector (Agilent 6890N, MSD 5975B, USA) using a 5 MS column (Agilent, USA). The temperatures of the detector and injector were 250°C and 220°C, respectively. The initial and final temperatures of the oven were 70°C and 250°C at a rate of 7°C/min. Helium was used as the carrier gas and the mass scan range was from 50 to 450 z/m at 70 eV. Identification was made using the NIST05 Mass Spectral library.
Determination of total and fecal coliforms
The Most Probable Number (MPN) of total coliforms, fecal coliforms, and Escherichia coli was obtained for the three sampling points (A1-A3) and followed the Mexican Norm NMX-AA-42-1987, which agrees with the ISO norm: ISO/DP 9308/2.
Biochemical Oxygen Demand (BOD)
The biochemical oxygen demand for five days for the three sampling points (A1-A3) was obtained with a Hach BOD Track II Respirometric apparatus according to manufacturers´ instructions.
Determination of organic carbon
The organic carbon content was evaluated in water samples for the sampling point A1 using a TOC-LSCH analyzer (Shimadzu, Japan) which principle of detection method is the combustion catalytic oxidation at 680 ºC. Total and inorganic carbon (TC and IC) were measured automatically in triplicates by the equipment and organic carbon was calculated by the difference between those values. The detection limit of TC and IC was 4 μg/L with a maximum coefficient of variation of 1.5% between replicates.
DNA extraction and PCR
Microbial community DNA was extracted directly from the slurries (A1) collected on January 21, 2013, using the UltraClean Soil DNA kit (MoBio Laboratories, Solana Beach CA) and PCR amplification of 16S rDNA genes was performed with the following primer pair: 63F (CAG GCC TAA CAC ATG CAA GTC) and 1389R (ACG GGC GGT GTG TAC AAG) (Marchesi et al., 1998) for bacteria as described previously (Aburto et al., 2009). The cycling conditions were as follows: 1 cycle at 94 °C for 2 min, 30 cycles of 1 min at 94 °C, 1 min at 55°C and 2 min at 72 °C and a final elongation at 72 °C for 10 min.
PCR amplification of genes encoding Shiga toxins 1 and 2 was performed on the A1 slurries with the primer pair stx1 and stx2 as described previously (Fagan et al., 1999). The cycling conditions were as follows: an initial 95°C denaturation step for 3 min followed by 35 cycles of 95°C for 20 s, 58°C for 40 s, and 72°C for 90 s. The final cycle was followed by a 72°C incubation for 5 min.
Cloning and sequencing
PCR products were cloned with a CloneJet cloning kit (Thermo) as described in the manufacturer´s instructions using One Shot TOP10 chemically competent E. coli cells. Recombinant colonies were recovered from LB agar plates containing ampicillin (50 mg ml-1). The screening of inserts from transformants was performed by direct PCR amplification from white colonies using the 16S primers 63F (CAG GCC TAA CAC ATG CAA GTC) and 1389R (ACG GGC GGT GTG TAC AAG). Heating at 94°C for 10 min preceded standard cycling conditions (as above). Amplified inserts were grouped on the basis of restriction fragment length polymorphism (RFLP) patterns using a combination of the two restriction endonucleases CfoI and AluI (Sigma) at 37°C for 3 hrs. Representative clones of each RFLP group were sequenced. The sequencing analysis was performed at the UNAM biotechnology institute (Mexico). The sequences length was 600 to 700 nucleotides in average.
Phylogenetic analyses
The sequences obtained were compared with the European Bioinformatics Institute and Genebank databases by online FastA searches (Pearson & Lipman, 1988) and to the Check Chimera program (Cole et al., 2003). The profile alignment technique of ClustalW (Thompson, Higgins, & Gibson, 1994) was used to obtain preliminary alignments that were manually refined in GeneDoc Multiple Sequence Alignment Editor ver. 2.6.002 (Nicholas & Nicholas, 1997) taking into account of secondary structure. Phylogenetic analyses were performed with the Phylip software package (Felsenstein, 2001), using the Jukes-Cantor distance and the neighbour-joining methods. The significance of the branching order was determined by bootstrap analysis with 1000 resampled data sets.
Results and discussion
Water quality
Total coliforms, fecal coliforms, and biochemical oxygen demand (BOD) values are shown in table 3. These values indicate that the waters are heavily contaminated according to the Mexican maximum limits (NOM-003-Semarnat-1997).
Table 3 MPN of total and fecal coliforms and BOD for sampling points A1-A3. A: Atoyac River.
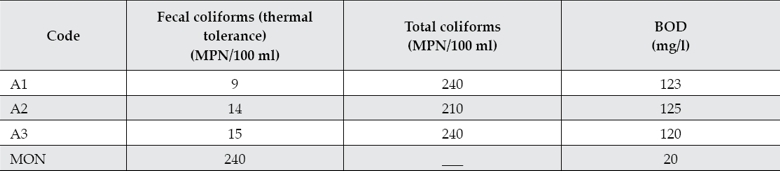
MON: Maximum limits of Mexican Official Norm (NOM-003-Semarnat-1997).
Metals, semimetals and other elements in sediments
Element concentrations for As, Ba, Ca, Cu, Fe, K, Mn, Mo, Pb, Rb, Sr, Ti, Zn, and Zr were obtained for sampling point A1 (table 4). The concentrations of As, Mo and Pb were below the detection level. The mean of values for Fe, Mn, Pb, and Zn detected in this study are below the permissible limits of the USEPA (1977) and the Ontario Ministry of the Environment and Energy (OME, 1976) except for Cu, which is slightly above the USEPA limit. They are also below the values determined for lead and arsenic in river sediments collected near the river origin in the state of Tlaxcala (García-Nieto et al., 2011) but above for Fe and Mn in agricultural soils located further south Puebla city that have been irrigated with the river waters for more than 30 years (Méndez-García, Rodríguez-Domínguez, & Palacios-Mayorga, 2000). This suggests that heavy metal concentrations in sediments vary along the river course and at the dam. Another study has also determined low concentrations of metals in the river waters (Cedeño, Téllez, Pacheco, Rosano, & Ascencio, 2008) and the water hyacinth (Eichhornia crassipes) has been suggested as a phytoaccumulation species within the Valsequillo dam (Cedeño et al., 2008). However, it has also been established that the concentrations of metals have accumulated in soils throughout the time because of the irrigation activities (Méndez-García et al., 2000) requiring constant monitoring. Moreover, further studies have established that these waters should not be used for the irrigation of crops (Díaz et al., 2005; Bonilla, Vázquez, Silva, & Cabrera, 2013) and this is supported by a previous study where long-term irrigation of soils with wastewater in Germany caused significant heavy metal contamination and a potential impact on the underlying aquifers (Lottermoser, 2012).
Table 4 The content of metals, semimetals and other elements in the Atoyac River sediments and comparison with the USEPA limits.
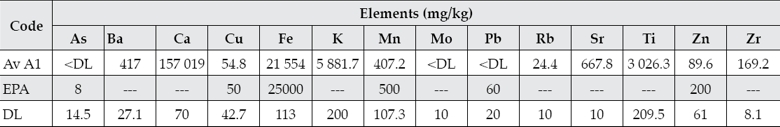
Av: Average value; A: Atoyac River; EPA: USEPA Guideline (1977); DL: Detection limit.
As, Mo and Pb were below the detection limit.
Organic compounds detection
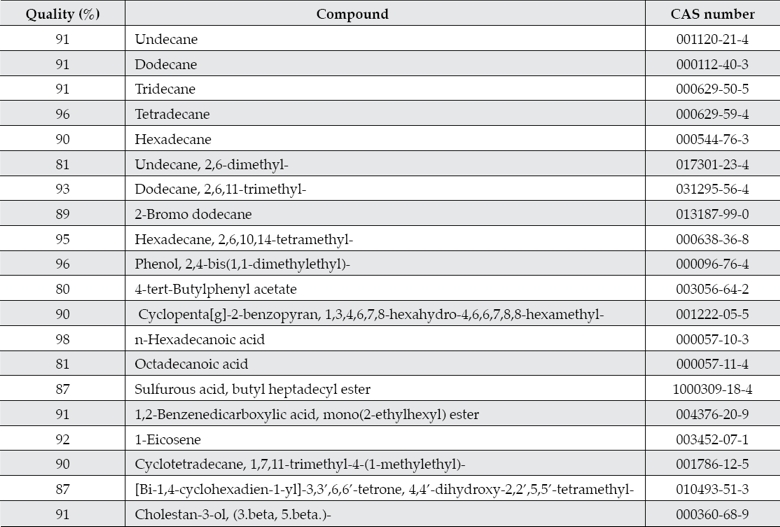
A total of 20 organic compounds were detected in the river sediments of sampling point A1 and they are higher alkanes (C11-C16), esters, organic acids, phenols, phthalates, among others (table 5). The detection quality of most of the compounds ranged from 80 to 98% (table 5). These compounds have been reported as common contaminants of river waters (Pennell, Abriola, & Weber, 1993; Gracia, Cortés, Sarasa, Ormad, & Ovelleiro, 2000). Ten of the twenty organic compounds detected in the sediment sample are classified by the USEPA as chemicals for potential endocrine disruptor screening and testing and they include n-hexadecanoic acid, octadecanoic acid, phenol, 2,4-bis(1,1-dimeth-ylethyl)-, dodecane, cholestan-3-ol, (3.beta,5.beta.)-, hexadecane, tridecane, undecane, tetradecane and Cyclopenta[g]-2-benzopyran, 1,3,4,6,7,8-hexahydro-4,6,6,7,8,8-hexamethyl(USEPA, 2012). Compounds such as cholestan-3-ol, (3.beta, 5.beta.), known also as coprostanol, have been used as indicators in faecal pollution source tracking (FST) to identify the origin(s) of pollution in water, in areas where diffuse faecal contamination are often marked by the co-existence of human and animal sources (Roslev & Bukh, 2011; Derrien et al., 2012). The presence of coprostanol was correlated with the microbial populations found in situ, as discussed later. Moreover, a recent study detected other persistent organic pollutants (POPs) in the river sediments that included hexachlorocyclohexane isomers, hexachlorobenzene, many polychlorinated biphenyl congeners and plaguicides such as DDT, DDE, mirex, and aldrin, (Juárez-Santacruz et al., 2012). Therefore, the presence of all these organic pollutants could pose a health risk to the population because the crops in the sub-basin and those located lands above the dam are irrigated with these waters. Crops that are irrigated with polluted waters may contain pesticide residues; chlorpyrifos, cypermethrin, and omethoate have been usually detected in green vegetables such as spinach (Kobayashi et al., 2011).
Bacterial diversity in situ
A total of 53 clones were retrieved from the clone library and they were related to members of the Enterobacteriaceae within the Gammaproteobacteria (figure 2). The bacterial diversity of the surveyed sediments is low; species related to Shigella flexneri, Escherichia fergusonii and E. coli dominated the clone library. The degradation of hydrocarbons has been reported recently by Escherichia fergusonii (Sriram et al., 2011; Pasumarthi, Chandrasekaran, & Mutnuri, 2013) suggesting the use of some of the detected compounds by the Escherichia fergusonii-related bacteria, however, these and the other members of the Enterobacteriaceae may also pose potential health risks to the population. Shigella flexneri (B serogroup) is a common cause of bacillary dysentery in humans. Infection by Shigella is usually via ingestion invading the colonic mucosa and producing an intense inflammatory reaction leading to tissue destruction (Kotloff et al., 1999). Therefore, we tested for the presence of Shiga genes within our samples but they were not found. However, future studies would be necessary to look for further genes encoding other toxins or virulence and invasion plasmids (Sansonetti, Kopecko, & Formal, 1982; Ménard, Sansonetti, & Parsot, 1993; Fagan et al., 1999) in this strain in order to have a proper assessment of the Shigella-related clones pathogenicity.
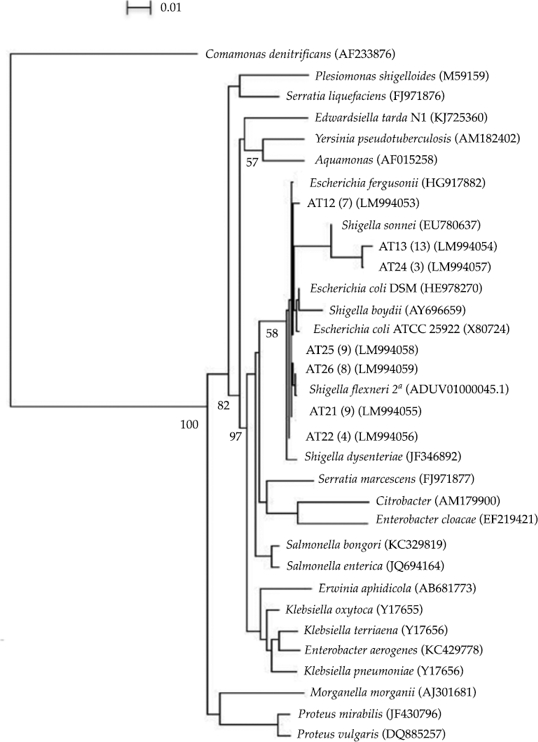
Figure 2 Phylogenetic tree of the Enterobacteriales order based on partial 16SrDNA sequences showing the relationships among bacterial clone sequences from this study and members of the Enterobacteriaceae family found in the database. Jukes and Cantor distance, neighbour-joining method, outgroup: Comamonas denitrifications. Clones from this study are in bold, A: Atoyac. A number in parenthesis indicates the number of clones obtained for each sequence. Accession numbers of the sequences retrieved from the databases are also indicated in parenthesis. Bootstrap values above 50% are shown at branch nodes (per 1000 trials). 16S rRNA sequence similarity over the region 800 to 1390 based on E. coli numbering.
The high number of clones related to Enterobacteriaceae is not uncommon since the river receives industrial, agricultural and domestic effluents; however, our samples were collected after the wastewater treatment process suggesting the presence of further domestic effluents or the lack of efficacy during the wastewater treatment process. This is also constant with the high levels of coliforms (240 MPN 100 ml-1) found at the same sampling point and with previous studies (Silva, Muñoz, Isla de Bauer, & Infante, 2002; Cabrera, Bonilla, Tornero, & Castro, 2005; Sandoval-Villasana et al., 2009). Thus, there may be health risks since these waters are later used to irrigate several crops such as maize, sorghum, and alfalfa in lands above the dam (Larenas-Bazán, 2010). Therefore, it is of great importance to identify the effluents located after the wastewater treatment process and/or review the water quality coming out of those treatment facilities, another aid would be the use of microbial source tracking markers such as the stanols (Derrien et al., 2012) in order to identify faecal contamination sources. It is equally important to look for catabolic and pathogenicity genes in the environmental samples in order to properly assess the microbial diversity degrading potential and its pathogenicity.
Conclusion
This study provides further evidence of the environmental degradation of the Atoyac River. It is the first to describe the bacterial communities in its sediments and to elucidate the contaminants present just prior to the dam. Moreover, it shows a low bacterial diversity in situ, usually related to gut diseases and although it lacks Shiga toxins encoding genes, further studies are required to discard the presence of all other toxin-encoding genes since these waters are later used for crops irrigation.











 nueva página del texto (beta)
nueva página del texto (beta)