Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias forestales
versión impresa ISSN 2007-1132
Rev. mex. de cienc. forestales vol.12 no.65 México may./jun. 2021 Epub 30-Ago-2021
https://doi.org/10.29298/rmcf.v12i65.781
Scientific article
Overexpression of PtCSP4 in poplar promotes greater potential for PCB phytoremediation
1Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias, Centro de Investigación Regional Pacífico Centro. Campo Experimental Tecomán. México.
Phytoremediation is a widely accepted biotechnology that reduces, absorbs and degrades toxic pollutants. Polychlorinated biphenyls (PCB) are persist organic pollutants that cause harmful effects to the environment. It has been acknowledged that poplar (Populus trichocarpa) is fast growing, it has high transpiration and a complete genome sequence. Thus, it is considered a perfect model in phytoremediation. Genetically modified plants can become more efficient remediation sources of soil and water contaminated sites with polychlorinated biphenyls (PCB). Yb-1 is a multifunctional protein involved in the regulation of transcription, translation, mARN splicing and ADN repair. Yb1 transcription factor, is known in humans and mammals but very little in plants. Yb-1 in plants is best known as CSP (cold shock proteins). In this work, PtCSP4 (Potri.004G172600) gene was isolated and amplified from poplar. The gateway recombination system was used to clone PtCSP4 in a binary vector with a constitutive promoter. PtCSP4 was introduced in Arabidopsis thaliana and its tolerance was tested in presence of PCB (Aroclor 1221). Eight transgenic lines of in vitro cultured Arabidopsis were exposed to PCBs. All results suggest that PtCSP4 is a good candidate for phytoremediation of PCB.
Key words: Poplar; Arabidopsis; aroclor; in vitro; PtCSP4; Populus trichocarpa Torr. A. & Gray ex Hook.; tolerance
La fitorremediación es un método biotecnológico ampliamente aceptado que reduce, absorbe y degrada los contaminantes tóxicos. Los bifenilos policlorados (PCB) son contaminantes orgánicos persistentes que causan efectos nocivos para el medio ambiente. Se reconoce que el álamo (Populus trichocarpa) tiene un crecimiento rápido, alta transpiración y se tiene una secuencia completa de su genoma; por lo que se considera un modelo perfecto en fitorremediación. Las plantas genéticamente modificadas pueden ser más eficientes en la remediación de sitios, cuyos suelo y agua están contaminados con bifenilos policlorados (PCB). Yb-1 es una proteína multifuncional involucrada en la regulación de la transcripción, traducción, empalme de ARNm y reparación de ADN. El factor de transcripción Yb1 se conoce en humanos y mamíferos, pero muy poco en plantas. Yb-1 en plantas es más conocido como CSP (proteínas de choque frío). En este trabajo, el gen PtCSP4 (Potri.004G172600) se aisló y amplificó a partir del álamo. El sistema de recombinación de puerta de enlace se usó para clonar PtCSP4 en un vector binario con un promotor constitutivo. PtCSP4 se introdujo en Arabidopsis thaliana y se probó su tolerancia en presencia de PCB (Aroclor 1221). Ocho líneas transgenicas de Arabidopsis cultivadas in vitro fueron expuestas a PCB. Todos los resultados sugieren que PtCSP4 es un buen candidato para la fitorremediación de PCB.
Palabras clave: Álamo; Arabidopsis; aroclor; PtCSP4; Populus trichocarpa Torr. A. & Gray ex Hook.; tolerancia
Introduction
Persistent Organic Pollutants (POPs) are particularly toxic and stable compounds, that include insecticides, herbicides, polychlorinated biphenyls (PCBs), dioxins and flame retardants (www.pops.int). These compounds are characterized by their great chemical stability, which has contributed, to a great extent, to their dispersion throughout the planet's ecosystems. Due to their stability and low solubility in water, PCBs are extremely difficult to remove from soil matrices. They were introduced as commercial blends, the most common of which was the Aroclor series (Produced by Monsanto Chemical Industry, Illinois) (Beyer and Biziuk, 2009; EPA, 2014). The amount of PCB-contaminated soils in the world is unknown. In the United States of America, 350 of the 1 290 contaminated sites included in the “Superfund” program of the Environmental Protection Agency (EPA) are contaminated with PCBs (27 %) (http: //www3.epa.gov/). As if that were not enough, due to their derived capacity to bioaccumulate, their physical stability and hydrophobicity, PCBs are present throughout the entire food chain of the planet (Malisch and Kotz, 2014).
The use of efficient remediation technologies is very important to try to tackle the problem of PCBs. One of these technologies is phytoremediation, which is based on the use of plants to extract, sequester and detoxify soil pollutants. This technology significantly reduces the costs of traditional decontamination (Gomes et al., 2013), while contributing to the fight against erosion and helping to restore degraded spaces (Dickinson et al., 2009). Identifying genes in plants that are involved in PCB metabolism can increase the effectiveness of phytoremediation. The ability of plants to degrade xenobiotics was discovered in 1940, in connection with the decontamination of pesticides (Sanderman, 1994). Since then, the development of molecular biology has contributed to elucidate the metabolism of some pollutants in plants (Eapen et al., 2007; Van Aken et al., 2009; Wang et al., 2012).
The pollutant degradation process can be summarized in three stages: transformation of the compound, conjugation and compartmentalisation. As part of the last stage, a subclass of ABC transporters, called multidrug resistance (MDR), are involved in the regulation of the influx and cellular exit of xenobiotics (Rea, 2007; Xi et al., 2011). Surprisingly, very little is known about the role of plant MDRs in xenobiotic resistance, as few studies have been performed (Conte and Lloyd, 2011). The mechanisms that govern these biochemical processes are finely regulated by a large number of genes that encode regulatory proteins. Transcription factors play a central role, altering the expression of numerous genes. Many of these factors are co-induced in response to different stresses, which suggests the existence of complex interactions (Yuan and Perry, 2011; Golldack et al, 2014). Campos (2011) analyzed the possible involvement of cold shock proteins (CSP) in response to treatments with organic pollutants. The CSP family in plants has a highly conserved domain, known as the cold shock domain (CSD), and has two or more zinc finger Zinc Knuckle domains of the CCHC type (CX2CX4HX4C), two RNA recognition sites (RRM) and repeat RGG boxes.
Some transgenic plant varieties have been generated for xenobiotic phytoremediation (Maestri and Marmiroli, 2011). Although much of the genetic improvement has been performed in laboratory model plants, such as Nicotiana tabacum L. and Arabidopsis thaliana (L.) Heynh, their low biomass and short life span, might not make them suitable for in situ remediation. Therefore, there is a particular interest in the genetic improvement of trees that maintain fast growth, extensive root systems and abundant biomass production, such as poplars and willows (Doty, 2015).
The Populus genus comprises 40 tree species native to the temperate and cold northern regions. It has high growth rates (up to 3 m yr-1), as well as a large root system that ensures efficient groundwater absorption (Isebrands and Richardson, 2015). The suitability of poplar does not only lie in its physiological characteristics. At present, Populus is the best available forest model system (genome sequenced). Therefore, poplar can be used as a model species for studies in molecular eco-physiology, as well as for gene-function associations in other trees (Brunner and Nilson, 2004).
The objective of this work was to demonstrate that transgenic lines of A. thaliana that overexpress PtCSP4 from poplar (Potri.004g172600) tolerate the presence of PCBs.
Materials and Methods
The study was carried out in the Forest Biotechnology Laboratory of the Center for Plant Biotechnology and Genomics (CBGP), located in the Science and Technology Park, UPM Campus de Montegancedo, Ctra, M-40, Km 38, 28223, Madrid, Spain.
Vegetal material
In vitro poplar plants (clone INRA 7171-B4) of Populus tremula × alba L. were used in the experiments. This clone was chosen for its excellent in vitro and ex vitro behavior, as well as its ease of being genetically transformed, in addition to its great vigor in the field. The Arabidopsis thaliana Columbia ecotype was used for genetic transformation. The seeds were obtained from the Arabidopsis Biological Resource Center in Ohio, USA. For the transformation of A. thaliana, the C58C1 (pMP90) strain of Agrobacterium tumefaciens (Smith & Townsend, 1907) Conn, 1942 was used, which contains the plasmid pMP90 derived from pTiC58. This strain was provided by Dr. Pamela Green (Michigan State University).
Treatments with Aroclor 1221 (PCB)
For the treatments with this xenobiotics, the growth of the poplars in vitro was previously assessed in the presence of different Aroclor 1221 concentrations (PCB mix commercialized by Sigma Aldrich®) diluted in sterile DMSO: 0 (control), 50 and 200 mg L-1. These experiments were carried out under in vitro conditions, using in each jar 20 mL of MS-phytoagar supplemented with DMSO (controls) or Aroclor 1221 diluted in the same volume of DMSO, with 1-month-old poplar trees that were transplanted to the contaminated medium. After determining the effect of PCBs on vegetative growth, the foliar biomass of 20 plants was monitored with a digital scale (KERN® Mod. PCB2500-2) after oven-drying them (ECOSHEL® Mod. 9023AV) at 70 °C for 24 h. Additionally, the root length from the roots measured with a stainless steel strip (MILLER® de 6”).
Genomic DNA extraction from P. tremula × P. alba
A total of 100 mg of whole poplar plant tissue treated with Aroclor 1221 was pulverized in liquid nitrogen. All reagents used herein were Sigma-Aldrich® unless differently stated. DNA was extracted using the DNeasy Plant Mini kit (Qiagen ®; Hilden, Germany), following manufacturers instructions. DNA was quantified with a NanoDrop® ND-100 spectrophotometer (NanoDrop Technologies; Wilmington, DE, USA) with a ratio of 280/260 ≥ 1.8. To visualize the DNA, agarose gels at 0.8 % (p/v) were used, prepared with TAE buffer (40 mM Tris HCl, pH 8.0, 20 mM sodium acetate, 1 mM EDTA) supplemented with ethidium bromide at 0.1 mg L-1. The samples were mixed with loading buffer (bromophenol blue 0.05 % (p/v) and glycerol 5 % (p/v)). DNA was visualized on a Molecular Imager Gel Doc XR System. For virtual quantification, Bio Rad® Quantity One 4.6.5 software was used (California, USA).
Extraction and sequencing of E. coli plasmid DNA
Plasmid isolation was carried out with the QIAprep Spin Miniprep kit (Qiagen®; Hilden, Germany). To check the quality, plasmid DNA was quantified with a NanoDrop® ND-100 spectrophotometer with a ratio of 280/260 ≥ 1.8. Restriction enzymes NdeI y HpaI (Takara Bio Inc®; Shiga, Japan) were used to check the insert size. The fragments were visualized on 0.8 % agarose gels. Sequencing was carried out at the National Cancer Research Center (CNIO) in Madrid, Spain, which has an ABI 3730 system® (Applied Biosystems; Foster City, CA, USA).
Heterologous expression of PtCSP4 in A. thaliana
The coding sequence of PtCSP4 (Potri.004G172600.1) was amplified by PCR from genomic DNA; entire aatb sites were introduced using the high fidelity Long Range DNA Polymerase enzyme (Takara Bio Inc®; Shiga, Japan). The fragment obtained (624 bp) was cloned using GatewayTM Invitrogen ® (Gaithersburg, MD, USA). The oligonucleotides pairs for cloning were designed from the genomic sequences, with a melting temperature of 60 °C. To clone the PtCSP gene, forward 5'attbAGAAGGAGAAAACGGCATGGGTGAGAGG 3' and reverse 5'attbCCTCACTAAAATCAACGTCC 3' oligonucleotides pairs were used, which were designed in the most divergent area of the 5' end of the coding region of the amino-terminal. Thermocycling conditions were initial 2 min at 50 ºC, 10 min at 95 ºC denaturation, followed by 40 amplification cycles of 15 sec at 95 ºC, 1 min at 60 ºC, to ensure the amplification of a single product. The PCR product, was cloned into the expression vector pDONR 221 GatewayTM (Invitrogen®) and later, in order for the transgene to remain under the control of the 35S promoter, it was transferred to the binary vector PB2GW7 GatewayTM (Invitrogen®). With the resulting construct, cells of A. tumefaciens strain C58C1 (pMP90) were transformed, which was used to infiltrate Arabidopsis plants (Figure 1). As a control, Arabidopsis plants were transformed with an empty vector without insert.
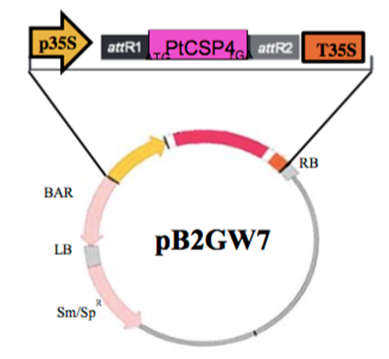
Figure 1 Cloning of the coding region of PtCSP4 in the vector pB2GW7. Cloning design for the ectopic expression of PtCSP4 in Arabidopsis thaliana under the control of the CaMV35S promoter. The region between the LB and RB (T-DNA) regions includes the BAR gene, which confers resistance to the herbicide ammonium glufosinate.
Cell Preparation and Transformation with Agrobacterium
The preparation of competent cells and their transformation was carried out as described by Hofgen and Willmitzer (1988), with some modifications. From an individualized colony, a culture was prepared in 10 mL of LB medium with rifampicin (50 mg L-1), gentamicin (25 mg L-1) and kanamycin (50 mg L-1). The culture was incubated at 28 °C and 200 rpm for 16 h. The following day, 1 mL was inoculated into 100 mL of the same medium (1: 100 dilution), incubated at 28 °C and 200 rpm until an OD600 ~ 0.8 was reached. Subsequently, the culture was centrifuged at 5 000 rpm and 4 °C for 5 min, the supernatant was discarded and the cells were washed with 20 mL of cold 150 mM NaCl (sterile). The cells were centrifuged again at 4 °C and 5 000 rpm for 5 min. and resuspended in 2 mL of 20 mM CaCl2 (sterile and cold).
For the transformation of A. tumefaciens, 1 ng of plasmid pB2GW7 was added to 10 µL of competent cells (Agro 58C1). They were incubated 5 min at 37 °C with gentle shaking, after which they were kept on ice for 30 min. Subsequently, 500 μL of LB medium were added and incubated for 3 h at 28 °C and 150 rpm (TECNAL®; Santo Domingo, Dominican Republic). After a 1 min centrifugation at low speed (2 000 rpm), the cell pellet was resuspended in 150 mL of LB medium and plated on LB-agar supplemented with rifampicin (50 mg L-1) and spectinomycin (100 mg L-1). The colonies were visible after incubating two days at 28 °C.
Genetic transformation of Arabidopsis thaliana
Three-week-old A. thaliana plants grown in 5 × 5 cm alveoli (9 seeds/alveolus), with 10 cm long inflorescences, were used. The transformation was carried out following the vacuum infiltration method described by Clough and Bent (1998), starting from appropriate A. tumefaciens cultures. Recombinant cells of strain C58C1 (pMP90), transformed with pB2GW7, were inoculated on LB plates supplemented with rifampicin (50 mg L-1) and spectinomycin (50 mg L-1). After two days shaking (200 rpm) at 28 °C, individual colonies were selected and pre-inocula were prepared in 3 mL of LB medium with the same antibiotics used for the solid medium. From these, 200 mL of LB medium were inoculated with antibiotics, maintaining the culture under the same conditions as before, until reaching an OD600 of 1.2. The cells were isolated by centrifugation (15 min at 4 °C and 5 500 rpm in a Beckman ® JA20 rotor) and were suspended in infiltration medium (MS salts 1 × pH 5.7; Sucrose 5 % (p/v), MES 0.05 % (p/v), 0.044 µM benzylaminopurine (BAP), Silwet ® L 77 0.02 % (v/v)) (Lehle Seeds), which is added immediately before infiltrating.
The infiltrations were carried out for 15 min under vacuum and with two repetitions. After drying the plants with absorbent paper, they were placed back in the culture chambers under normal growing conditions. The transformed lines were selected by sowing the seeds in MS medium supplemented with the herbicide BASTA (phosphinothricin) (10 µg mL-1). After two weeks, the live Arabidopsis plants were transferred to a new herbicide plate to rule out false positives. For the analysis, the T3 plants were obtained. The success of the transformation was verified by qRT-PCR amplifications.
Total RNA extraction of P. tremula × P. alba for real-time PCR validation
RNA extraction was performed using 200 mg of pulverized tissue (from complete in vitro plant). For initial extraction, the phenol-chloroform method (Ambion, 2008) was used and subsequently purified with the RNeasy Plus Mini kit (Qiagen ® ; Hilden, Germany). Finally, it was eluted in 60 µL of sterile RNase-free water. RNA was quantified in a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies) with an absorbance ratio of 260 nm/280 nm ≥ 2. With this extraction methodology, 200 ng of total RNA was obtained. To validate RNA quality and quantity, an aliquot was sent to the National Center for Oncological Research (CNIO) for nanocapilarity electrophoresis with the Agilen Bioanalizer 2100 (Agilent Technologies, Inc.; Santa Clara, CA, USA).
Before performing cDNA synthesis, the RNA was treated with DNases to avoid contamination with genomic DNA in the samples. The Turbo DNA-Free kit (Ambion, Applied Biosystems®; Austin, TX, USA) was used for this purpose. The cDNA was synthesized from 1.5 µg of RNA in 30 µL of reaction of the PrimeScriptTM RT reagent kit (Perfect Real Time) (Takara Bio, Inc®; Otsu, Japan) in the presence of the primers "Oligo dT" and "Random 6 mers", in the amounts recommended by the manufacturer, with an incubation of 90 minutes at 37 °C.
Quantitative real-time PCR (qRT-PCR)
The recent real-time PCRs were carried out on an ABI PRISM 7300 thermal cycler (Applied Biosystems; Foster, CA, USA). All reactions were performed in a final volume of 20 µL, with 10 µL of the SYBR™ Green 2x mixture (Applied Biosystems; Foster, CA, USA), 0.2 µM of each oligonucleotide, and 1 µL of cDNA (section 3.7.2.). Ribosomal RNA subunit (18s) and polyphenol oxidase (PPO) genes were used as reference genes; cDNA was used at a 1:100 dilution. The amplification cycles were an initial step of 2 min at 50 °C, a denaturation step of 10 min at 95 °C and for 40 amplification cycles of 15 sec at 95 °C and 1 min at 60 °C. Data were analyzed using 7300 Real-Time PCR System software. For the analysis of the expression levels, a relative quantification of the gene under study was made, by means of the Ct (2-∆∆Ct) method exposed by Livak and Schmittgen (2001).
Results and Discussion
Vegetative growth in the presence of Aroclor 1221
To elucidate the mechanisms that are activated on contact with PCBs, untreated poplar plants (control) were compared with plants treated with Aroclor 1221. Treatment conditions were previously optimized, and the symptoms resulting from exposure to PCB were evaluated. One-month old in vitro rooted poplars were used in the analysis in order to avoid gene expression associated with very young stages, and also to minimize the possible effects of transplants. In vitro cultivation of woody plants is generally considered very difficult, although there are more or less efficient regeneration protocols for some species. The hybrid chosen is particularly viable and, in fact, leads the literature on forest biotechnology (Harfouche et al., 2011; Isebrands and Richardson, 2015).
Comparisons between treated and control plants revealed significant differences in some quantitative variables. Even though the treated in vitro plants exhibited stress characteristics under the tested conditions, they did not die and continued growing. Overall, plant growth and root system decreased as PCB concentration and exposure time increased. Also, symptoms of leaf chlorosis and necrosis became apparent. Biomass reduction was more pronounced in the aerial part than in the roots. The mean length of the roots of the control plants after five days was 3.58 cm, and was significantly different to the treated plants (n=30, F= 4.0, p=0.68), which averaged 4.0 and 3.1 cm at 50 and 200 mg L-1 of PCB, respectively. At 15 days, a length of 2.54 cm was recorded at 50 mg L-1 and 1.3 cm at 200 mg L-1; at this time, at 200 mg L-1, root length was 25 % of that of the control (Figure 2A). The same pattern was observed in the evaluation of foliar dry weight (biomass), in which the amount of biomass was lower due to the decrease in growth, and a 30 % decrease was observed in treated plants when compared to the control (Figure 2B). Symptoms of chlorosis and foliar necrosis also appeared. These effects are consistent with typical cellular stress responses (Proudfoot et al., 2002; Yang et al., 2012). In the present study, the morphological and growth analysis have served to assess the response of poplar to different levels of PCBs and to select the optimal experimental conditions for the treatments.
Verification for heterologous expression of PtCSP4
The PCR products obtained were analyzed by electrophoresis, where the size of the expected fragment in each of the constructs cloned in the Gateway system, were verified. Results are shown in Figure 3. Juárez-Reyes (2007), performed the steps followed for the expression vectors using gel electrophoresis.
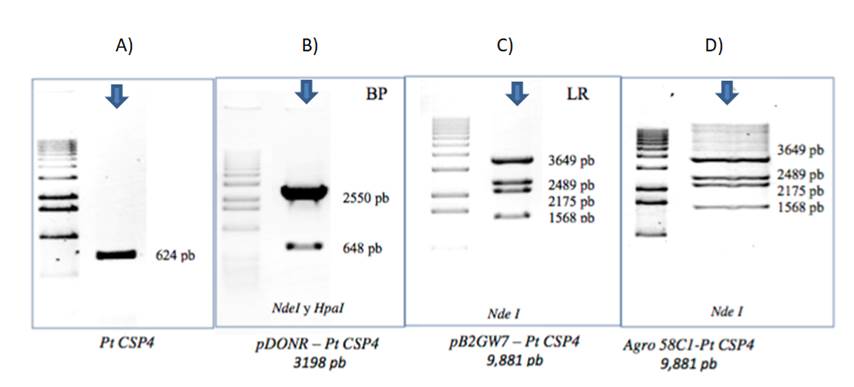
Figure 3 Verification of the cloning of PtCSP4 in agarose gels with restriction enzymes. The cloning product is marked with arrows. Checks for each cloning, A) The obtained PtCSP4 fragment (624 bp). B) Reaction LR was cloned into the expression vector pDONR-PtCSP4 of 3198 bp using the restriction enzymes NdeI and HpaI; C) the transgene was left under the control of the 35s promoter in a binary vector PB2GW7-PtCSP4 generating a 9881 bp fragment with the help of the restriction enzyme NdeI; D) resulting construct, cells of Agrobacterium strain C58C1 (pMP90) were transformed, which was used to infiltrate Arabidopsis plants.
Expression level of the transgene
Once stable transgenic lines were obtained, we proceeded to collect material from each line to analyze trangene expression. A total of eight independent transgenic lines were analyzed by qRT-PCR. Figure 4 presents average data from three technical replicates. Analogous results were reached in the two biological replicates. The relative abundances of the PtCSP4 transcript proved to be quite variable, as is often the case in related experiments. The signal recorded by the plants transformed with the empty plasmid was used to relativize the accumulation of transcript in the transgenic lines.
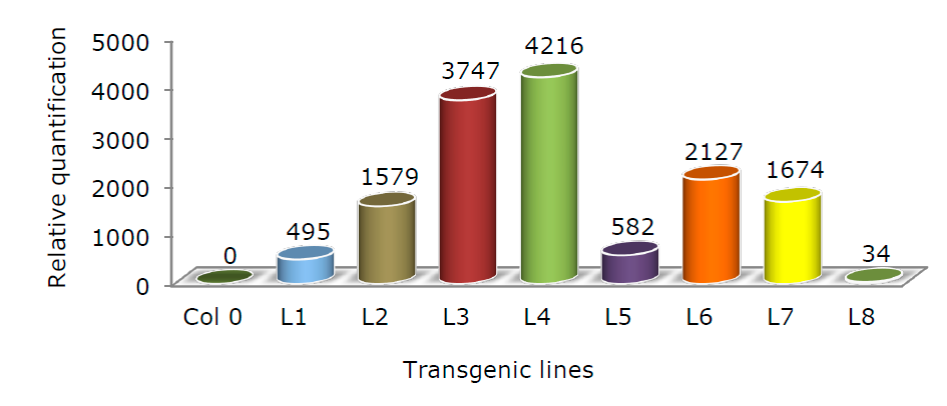
Figure 4 Expression of PtCSP4 in transgenic Arabidopsis plants. Average transcript levels detected in transgenic plants by qRT-PCR. The mean of 3 independent technical replicas is represented. The numerical values were normalized to the levels of an internal control (18S rRNA), and a value of 1 was assigned to the Col0 plants (Control).
As observed in Figure 4, the high expressing lines showed inductions 3 500 times greater (L3 and L4) than the control line; the lines with intermediate expressions increased between 1 500 and 2 000 times (L2, L6 and L7) and the low expressing ones below 500 times (L1, L5 and L8). From these data, lines L1, L4 and L6 were selected for the pollutant tolerance tests. It should be noted that no difference was observed in the growth and development of the plants as a consequence of the overexpression of PtCSP4.
Tolerance of transgenic lines to Aroclor 1221
Tolerance tests were carried out in the presence of PCBs with plants germinated and grown in vitro under controlled conditions. Until now, no data had been published on the tolerance of A. thaliana to Aroclor, so, before approaching these trials, the natural tolerance of Arabidopsis to this compound was analyzed.
The same doses used for poplar (50 to 200 mg L-1) were evaluated and it was found that Aroclor concentrations at 200 mg L-1 caused a clearly appreciable mortality. However, at concentrations lower than 100 mg L-1 there was no direct relationship between the presence of the compound and the symptoms observed in the seedlings, which is why 200 mg L-1 was used for tolerance tests. Once the test was optimized, the transgenic lines were subjected to the same treatment applied to untransformed plants. For this test, the plates were divided into four quadrants, 3 for transgenic lines and one for the control. Visual inspection of the results indicates that the lines that constitutively accumulate PtCSP4 have a greater tolerance to the presence of Aroclor at 200 mg L-1, a non-lethal concentration, but sufficient to create symptomatic stress in the plants (Figure 5).
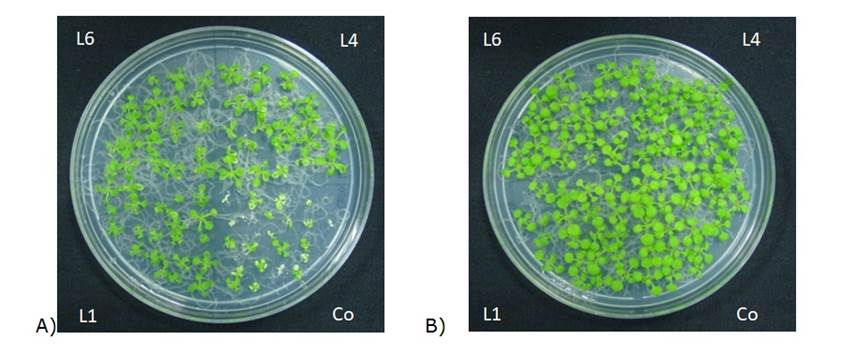
Figure 5 Transgenic lines with greater tolerance to the contaminant. A) Tolerance to 200 ppm of Aroclor 1221. B) Control plants.
When evaluating the plants ten days after germination under the contaminant treatment, the survival of the transgenic lines was 70 %, in contrast to 30 % of the control plants. In transgenic plants, the fresh weight value was 3 times higher than that of the controls, without significant differences between lines.
The efficacy of transgenic plants for the phytoremediation of organic pollutants has been proven in different species (James and Strand, 2009). The overexpression of the cotton (Gossypium arboretum L.) laccase gene (LAC1) in A. thaliana resulted in increased laccase-secreting activity and exhibited better resistance to trichlorophenol in soils. The control plants showed severe chlorosis, whereas LAC1 expressing plants showed less damage and better resistance to a variety of phenolic compounds. The data suggested that LAC1 was secreted from transgenic plants to metabolize trichlorophenol ex situ resulting in a less toxic environment (Wang et al., 2004).
In a similar study, laccase from Trametes versicolor (L.:Fr.) Quél (SIN: Coriolus versicolor) was expressed in tobacco, resulting in rhizosphere laccase secretion and increased degradation of bisphenol A and pentachlorophenol (PCP) in hydroponic cultivation (Sonoki et al., 2005). However, degradation in soils was not examined. Wevar et al., (2005) developed transgenic tomatoes (Lycopersicon esculentum Mill. cv. (Pear) that overexpressed TPX1, a native peroxidase that resulted in a higher peroxidase activity (Wevar et al., 2005). A manganese gene peroxidase (Mano) was expressed in tobacco plants (Iimura et al., 2002). Mano activity in a medium containing transgenic root cuttings was 50 times higher compared to controls, suggesting secretion of the MnP enzyme. A similar incubation in the presence of PCP resulted in an approximately 2-fold reduction in PCP concentration when comparing transgenic lines to controls.
The same research group transformed cottonwood (P. seiboldii × P. gradientata) with MnP from Trametes versicolor. Several transgenic lines had higher MnP activity than the control and contributed to a faster elimination of bisphenol A (Iimura et al., 2007). Given the above, it is very important to evaluate the biotechnological potential, as well as the overexpression of PtCSP. In later work, the necessary functionality tests will be carried out on the transgenic lines.
Differential expression of the PtCSP4 gene
Differential expression in the in vitro P. tremula × alba clones exposed to a concentration of 200 mg L-1 of Aroclor 1221 for 15 days, was validated through qRT-PCR (Figure 6). Chen et al., (2015) y Ariani et al., (2015) performed a validation of results through qPCR in P. tomentosa y P. × canadensis, respectively, revealed the empiric expression. It should be noted that this transcription factor had never been associated to xenobiotic stress before.
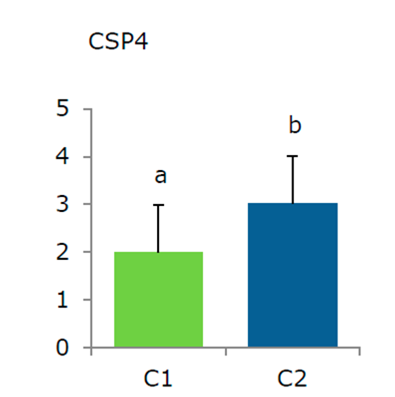
Figure 6 Empirical analysis of differential expression by qRT-PCR at 15 d exposure, C1: 50 mg L-1 and C2: 200 mg L-1. Changes in gene expression were estimated with respect to a control (fold change). The differential expression of cold shock (CSP4) Potri.004G172600.1 was validated. The bars indicate the standard error of the average fold change. Values were normalized according to 18s MRNA levels and level 1 was assigned to the value measured between controls.
It can be inferred that tolerance of the in vitro clones of P. tremula × alba results from PCSP4 expression levels and from a cascade of genes that participate in stress defense induced by PCB, or oxidative stress. However, it is important to mention that in order to obtain greater precision in to potential impact on gene expression, further transcriptomic and proteomic analysis must be performed.
Conclusions
It is not easy to obtain contaminant tolerant plants. Several CSP proteins are related to abiotic stress, among other processes such as growth, flowering, for example, but they have never been related to xenobiotic stress. Arabidopsis plants overexpressing poplar PCSP4 have shown greater tolerance to stress by PCBs (Aroclor 1221). This was evidenced from the decreased severity in the visible damage caused to the transgenic plants compared to the control. Over-expressing lines behave as if they had undergone acclimatization to stress. However, further in-depth characterization of its function and behavior in the field will be required for phytoremediation purposes, as well as proteomic and transcriptomic analysis.
Acknowledgements
To Conacyt for its support during the doctoral studies as well as to the Universidad Politécnica de Madrid, Spain; to INIFAP for its trust and support. Also, I would like thank the Centro de Biotecnología y Genómica de Plantas (CBGP) of the Universidad Politécnica de Madrid, with special regards to Dr. Luis Gómez Fernández and to Dr. Ángela Bibiana Contreras Mogollón.
REFERENCES
Ambion. 2008. The basics: RNA isolation. http://www.ambion.com/techlib/basics/rnaisol/index.html#1 (29 de agosto de 2016). [ Links ]
Ariani, A., D. Di Baccio, S. Romeo, L. Lombardi, A. Andreuccia, A. Lux, D. S. Horner and L. Sebastiani. 2015. RNA Sequencing of Populus × canadensis roots identifies key molecular mechanisms underlying physiological adaption to excess zinc. PLoS ONE 10(2): 1-20. Doi:10.1371/journal.pone.0117571. [ Links ]
Beyer, A. and M. Biziuk. 2009. Environmental fate and global distribution of polychlorinated biphenyls. Reviews of Environmental Contamination and Toxicology (201): 137-158. Doi: 10.1007/978-1-4419-0032-6_5. [ Links ]
Brunner, A. M. and O. Nilsson. 2004. Revisiting tree maturation and floral initiation in the poplar functional genomics era. New Phytologist 164(1): 43-51. Doi:10.1111/j.1469-8137.2004.01165.x. [ Links ]
Campos, V. M. 2011. Fitorremediación de contaminantes persistentes. Una aproximación biotecnológica utilizando chopo (Populus) como sistema modelo. Tesis de Doctoral. Facultad Montes. Universidad Politécnica de Madrid. Madrid, España. 196 p. [ Links ]
Chen, J., B. Chen and D. Zhang. 2015. Transcript profiling of Populus tomentosa genes in normal, tension, and opposite wood by RNA-seq. BMC Genomics 16: 164. Doi: 10.1186/s12864-015-1390-y. [ Links ]
Clough, S. J. and A. F. Bent. 1998. Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant JouARNl. 16(6):735-43. Doi:10.1046/j.1365-313x.1998.00343.x. [ Links ]
Conte, S. S. and A. M. Lloyd. 2011. Exploring multiple drug and herbicide resistance in plants- Spotlight on transporter proteins. Plant Science 180:196-203. Doi: 10.1016/j.plantsci.2010.10.015. [ Links ]
Dickinson, N. M., A. J. M. Baker, A. Doronila, S. Laidlaw and R. D. Reeves. 2009. Phytoremediation of inorganics: realism and synergies, InteARNtional JouARNl of Phytoremediation 11(2): 97-114. Doi: 10.1080/15226510802378368. [ Links ]
Doty, S. 2015. Improving crop growth, biomass production and phytoremediation using endophytes of poplar. Australasian Biotechnology (25): 46-63. https://search.informit.com.au/documentSummary;dn=295821182110914;res=I ELHEA>ISSN: 1036-7128 (15 de junio de 2018). [ Links ]
Eapen, S., S. Singh and S. F. D'Souza. 2007. Advances in development of transgenic plants for remediation of xenobiotic pollutants. Biotechnology Advances (25): 442-51. Doi:10.1016/j.biotechadv.2007.05.001. [ Links ]
Environmental Protection Agency of the United States (EPA). 2014. Evaluation of health hazards by exposure to Polychlorinated biphenyls (PCB) and proposal of a health-based quality criterion for soil. The Danish, Copenhagen, Denmark. http://www3.epa.gov/ (22 de enero de 2016). [ Links ]
Golldack, D., C. Li, H. Mohan and N. Probst. 2014. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Frontier in Plant Science (5): 147-151. Doi: 10.3389/fpls.2014.00151. [ Links ]
Gomes, H., C. Dias-Ferreira C. and A. B. Ribeiro. 2013. Overview of in situ and ex situ remediation technologies for PCB-contaminated soils and sediments and obstacles for full-scale application. Science of the Total Environment (446):237-260. Doi: 10.1016/j.scitotenv.2012.11.098. [ Links ]
Harfouche, A., R. Meilan and A. Altman. 2011. Tree genetic engineering and applications to sustainable forestry and biomass production. Trends in Biotechnology 29:9-17. Doi: 10.1016/j.tibtech.2010.09.003. [ Links ]
Hofgen, R. and L. Willmitzer. 1988. Storage of competent cells for Agrobacterium transformation, Nucleic Acids Research 20 (16): 9877-9877. Doi: 10.1093/nar/16.20.9877. [ Links ]
Iimura, Y., S. Ikeda, T. Sonoki, T. Hayakawa, S. Kajita, K. Kimbara, K. Tatsumi and Y. Katayama. 2002. Expression of a gene for MN-peroxidase from Coriolus versicolor in transgenic tobacco generates potential tools for phytoremediation. Applied microbiology and biotechnology. 59. 246-51. Doi: 10.1007/s00253-002-1008-6. [ Links ]
Iimura, Y., M. Yoshizumi, T. Sonoki, M. Uesugi, K. Tatsumi, K. Horiuchi, S. Kajita and Y. Katayama. 2007. Hybrid aspen with a transgene for fungal manganese peroxidase is a potential contributor to phytoremediation of the environment contaminated with bisphenol A. Journal of Wood Science. 53. 541-544. Doi: 10.1007/s10086-007-0890-z. [ Links ]
Isebrands, J. G. and J. Richardson. 2015. Poplars and willows: Trees for society and the environment. Environmental Forestry Consultants LLC, New London, WI, USA. pp.634. Doi: 10.1079/9781780641089.0000. [ Links ]
James, C. A. and S. E. Strand. 2009. Phytoremediation of small organic contaminants using transgenic plants. Current Opinion in Biotechnology. (20):237 -241. Doi: 10.1016/j.copbio.2009.02.014. [ Links ]
Juárez R., A. 2007.Delimitación de secuencias involucradas en el silenciamiento y transactivación de los genes tardíos del Virus Huasteco del Chile (PHV). Maestro en Ciencias en Biología Molecular. Instituto Potosino de Investigación Científica y Tecnológica A.C. (IPYCYT). San Luis Potosí, S.L.P., México. 67 p. [ Links ]
Livak, K. J. and T. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods (25): 402-408. Doi:10.1006/meth.2001.1262. [ Links ]
Maestri, E. and N. Marmiroli. 2011. Transgenic plants for phytoremediation. International Journal of Phytoremediation (13):264-79. Doi:10.1080/15226514.2011.568549. [ Links ]
Malisch, R. and A. Kotz. 2014. Dioxins and PCBs in feed and food-review from European perspective. Science of the Total Environment. 1:491-492. Doi: 10.1016/j.scitotenv.2014.03.022. [ Links ]
Proudfoot, N., A. Furger and M. Dye. 2002. Integrating mARN processing with transcription. Cell (108): 501-512. Doi: 10.1016/S0092-8674(02)00617-7. [ Links ]
Rea, P. A. 2007. Plant ATP binding cassette transporters. Annual Review of Plant Physiology 58: 347-375. Doi: 10.1146/annurev.arplant.57.032905.105406. [ Links ]
Sandermann, H. 1994. Higher plant metabolism of xenobiotics: the “green liver concept”. Pharmacogenetics 4(5): 225-241 Doi:10.1097/00008571-199410000-00001. [ Links ]
Sonoki, T., S. Kajita, S. Ikeda, M. Uesugi, K. Tatsumi, Y. Katayama and Y. Iimura. 2005. Transgenic tobacco expressing fungal laccase promotes the detoxification of environmental pollutants. Applied Microbiology and Biotechnology (67): 138-142. Doi:10.1007/s00253-004-1770-8. [ Links ]
Van Aken, B., P. A. Correa and J. L. Schnoor. 2009. Phytoremediation of polychlorinatedbiphenyls: new trends and promises. Technology (44):2767-2776. Doi:10.1021/es902514d. [ Links ]
Wang, G. D., Q. J. Li, B. Luo and X. Y. Chen. 2004. Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nature Biotechnology (22):893-897. Doi: 10.1038/nbt982. [ Links ]
Wang, Y., M. Qiao, Y. Liuand and Y. Zhu. 2012. Health risk assessment of heavy metals in soils and vegetables from wastewater irrigated area, Beijing-Tianjin city cluster, China. Journal of Environmental Sciences 24(4): 690-698. Doi:10.1016/s1001-0742(11)60833-4. [ Links ]
Wevar O., A. L., E. Agostini M., A. Talano, C. Capozucca, S. R. Milrad, H. A. Tigier and M. I. Medina. 2005. Overexpression of a basic peroxidase in transgenic tomato (Lycopersicon esculentum Mill. cv. Pera) hairy roots increases phytoremediation of phenol. Plant Sciences. (169): 1102- 1111. Doi:10.1016/j.plantsci.2005.07.007. [ Links ]
Xi, J., P. Xu and C. B. Xiang. 2011. Loss of AtPDR11, a plasma membrane- localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. The Plant JouARNl. 69: 782-791. Doi: 10.1111/j.1365-313X.2011.04830.x. [ Links ]
Yang, Z. B., D. Eticha, A. Albacete, I. M. Rao and T. Roitsch, 2012. Horst Physiological and molecular analysis of the interaction between aluminium toxicity and drought stress in common bean (Phaseolus vulgaris). Journal of Experimental Botany 63(8): 3109-3125. Doi: 10.1093/jxb/ers038. [ Links ]
Yuan, L. and S. E. Perry. 2011. Plant Transcription Factors: Methods and Protocols. Springer science+business media. New York, NY, USA. pp. 307-321. [ Links ]
Received: May 27, 2020; Accepted: August 13, 2020











 texto en
texto en 



