Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias pecuarias
versión On-line ISSN 2448-6698versión impresa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.14 no.4 Mérida oct./dic. 2023 Epub 17-Nov-2023
https://doi.org/10.22319/rmcp.v14i4.6354
Articles
Assessment of antibiotic resistance in fecal samples from calves with diarrhea in the Cajamarca region, Peru
a Instituto Nacional de Innovación Agraria (INIA). Estación Experimental Baños del Inca. Dirección de Desarrollo Tecnológico Agrario, Jr. s/n Wiracocha, Baños del Inca, Cajamarca 06004, Perú.
b INIA. Dirección de Desarrollo Tecnológico Agrario, La Molina. Lima. Perú.
Diarrhea is associated with infectious bacteria that cause mortality in calves, such as Escherichia coli, representing a problem for milk and meat producers globally, causing large economic losses. This study assessed the resistance to E. coli strains isolated from diarrheal feces of newborn calves from the Cajamarca region. Fifty two (52) fecal samples from calves from five provinces of the Cajamarca region were collected for the isolation of E. coli on MacConkey agar with sorbitol. The molecular identification of E. coli was performed by amplification of the uidA gene by conventional PCR and then antibiotic susceptibility/resistance was assessed using the Kirby-Bauer methodology and antibiotic discs with neomycin, tetracycline, sulfamethoxazole-trimethoprim and enrofloxacin. The results were that 96.15 % of E. coli strains were resistant to tetracycline, 51.92 % to sulfamethoprim, 26.92 % to neomycin and 9.61 % to enrofloxacin. It was also demonstrated that 30.76 % had resistance to two drugs, 19.23 % to three drugs and 5.76 % to four drugs; a significant difference was found in resistance to tetracycline (P<0.0001). It is concluded that newborn calves from the Cajamarca region that presented diarrhea are carriers of antibiotic-resistant E. coli, representing a problem for cattle farmers, since these strains can cause the death of animals and contribute to the spread of antibiotic resistance.
Keywords Resistance; Antibiotics; E. coli; Calves; Diarrhea
La diarrea está asociada con bacterias infecciosas que ocasionan mortalidad en terneros como Escherichia coli, representando un problema para los productores de leche y carne a nivel global, provocando grandes pérdidas económicas. En este estudio se evaluó la resistencia a cepas de E. coli aisladas de heces diarreicas de terneros recién nacidos de la región Cajamarca. Se recolectaron 52 muestras de heces de terneros de cinco provincias de la región Cajamarca para el aislamiento de E. coli en agar MacConkey con sorbitol. La identificación molecular de E. coli se realizó mediante la amplificación del gen uidA por PCR convencional y luego se evaluó la susceptibilidad/resistencia a antibióticos utilizando la metodología de Kirby-Bauer y el uso de discos de antibiótico con neomicina, tetraciclina, sulfametoxazol-trimetroprim y enrofloxacina. Los resultados fueron que el 96.15 % de cepas de E. coli fueron resistentes a tetraciclina, el 51.92 % a sulfametropim, el 26.92 % a neomicina y el 9.61 % a enrofloxacina. También se demostró que el 30.76 % presentaban resistencia a dos fármacos, el 19.23 % a tres fármacos y el 5.76 % a cuatro fármacos; se encontró diferencia significativa de resistencia a tetraciclina (P<0.0001). Se concluye que los terneros neonatos de la región Cajamarca que presentaban diarrea son portadores de E. coli resistentes a antibióticos, representando un problema para los criadores de ganado vacuno, ya que estas cepas pueden causar la muerte de los animales y contribuyen a la diseminación de la resistencia de antibióticos.
Palabras clave Resistencia; Antibióticos; E. coli; Terneros; Diarrea
Introduction
Diarrhea in calves has been linked to infectious pathogens, representing a challenge for those engaged in milk and meat production globally1. The most important infectious bacteria that cause mortality associated with diarrhea in calves are E. coli and Salmonella2, generating large economic losses if the disease is not treated in time with appropriate antimicrobials and supportive therapy2,3. Antibiotics have been used in animals for the treatment of diseases, prevention and control of common infections4,5, however, the inappropriate and excessive use of antibiotics contributes to antimicrobial resistance, threatening the health of animals and humans6. In relation to animals that are destined for slaughter and finally for human consumption, they act as reservoirs of antimicrobial-resistant strains7.
For example, it has been reported that, in Egyptian dairy farms, strains of E. coli have been isolated from diarrheal feces of calves, which were resistant to ampicillin8. Calves frequently eliminate microorganisms through their feces, generating the spread of bacteria in the farm environment, which could cause infections in other animals. E. coli isolates have been obtained from feces of dairy calves, which present resistance to multiple antibiotics from the fluoroquinolone group and the iucD gene was determined as the most prevalent9. Likewise, other studies mention the aerobactin operon (iucD), which produces four types of siderophores: enterobactin, salmochelin, aerobactin and yersiniabactin. The genes involved in the biosynthesis of siderophores are found in uropathogenic strains (UPEC) and commensal strains; nevertheless, salmochelin, aerobactin and yersiniabactin are located in UPEC-associated pathogenicity islands, but not in commensal strains, suggesting that iron uptake systems were acquired by horizontal gene transfer. Aerobactin is a siderophore present in most UPEC strains, having a great stability in the binding to Fe3+ and is one of the responsible for iron sequestration during a urinary tract infection (UTI), the combination of adhesion/iron uptake/toxicity genes shows the high virulence and the potential for damage that the strains have to cause a UTI9,10.
On the other hand, the French surveillance network for antimicrobial resistance in sick animals indicated that E. coli strains carrying most resistances have been isolated from diarrheal feces of neonatal calves10, with amoxicillin, tetracycline and streptomycin being the main antibiotics to which resistance has been generated7,11.
It is necessary to report the resistance that has been generated to multiple types of antibiotics to consider their control and proper use in cattle, because it represents a danger to public health. In addition, there are very few studies of antibiotic resistance in newborn cattle in Peru, with this study being a contribution to have knowledge of the current situation. The objective of the research was to assess the antibiotic resistance of E. coli strains isolated from diarrheal feces of newborn calves from five provinces of Cajamarca.
Material and methods
Study location
Work was carried out in 18 dairy farms under a semi-intensive system, which are located in the province of Chota, San Miguel, Celendín, San Marcos and Cajamarca, region of Cajamarca, Peru. A total of 52 samples were collected, which were from calves with diarrhea until the first month of life, of which 35 calves were males and 17 calves were females, in the rainy season (November 2020 - May 2021) (Figure 1). The fecal samples (approximately 3 g) were obtained directly from the rectum through the use of first-use sterile polyethylene bags, these samples were identified and taken in an expanded polystyrene box containing ice to the laboratory of Biotechnology in Animal Health| of the Baños del Inca Agricultural Experimental Station, where the isolation of E. coli from all the samples was carried out.
Isolation and identification of Escherichia coli
With a sterile bacteriological loop, 300 μl of feces was seeded on MacConkey II agar with sorbitol (Becton, Dickinson and Company® Loveton Circle Sparks, MD 21152, USA), then incubated at 37 °C for 24 h in an oven (Universal oven Memmert UN-110/ Schwabach/Germany), colonies with typical morphology and development, such as bright red colonies, were isolated, which were considered as E. coli.
Extraction of deoxyribonucleic acid (DNA) from Escherichia coli
Three selected E. coli colonies grown on MacConkey II agar with sorbitol were cultured in liquid microbial growth and multiplication medium 2xYT medium (Sigma, REF. Y2377) at 37 °C for 18 h; bacterial growth was determined by concentration of preset values in the spectrophotometer (PCR MAX Lambda 64272, Bibby Scientific Ltd. United Kingdom), the calculation of the Colony Forming Units (CFUs) was performed at 600 nm, determining the growth of viable bacteria in the growth medium. To obtain the E. coli DNA template, the Wizard® Genomic DNA purification kit (Promega, REF. A1120) was used, with the manufacturer’s indications, the DNA was stored in 1,500 μL polypropylene microtubes (Eppendorf™) in refrigeration, using a Samsung refrigerator, RT35K5930S8/PE, Samsung, Mexico of 4 °C, which is used for the different processes of the Polymerase Chain Reaction (PCR).
Molecular identification of Escherichia coli
The molecular identification of E. coli was performed by PCR using “primers” (F5’-TCAGCGCGAAGTCTTTATACC-3’, R5’-CGTCGGTAATCACCATTCCC-3’), for the amplification of the uidA gene (248 bp)11,12,13. In the PCR reaction, 1 μl (10 mM) of F and R of each primer, 7.7 μl of molecular grade water and 12.5 μl of G2 Green Master Mix (Promega, Madison, USA) were used, 2.8 μl of DNA at a concentration of 50 μg/ml was used as template. The thermal profile of the PCR reaction was: denaturation 94 °C/2 min, 25 cycles denaturation 94 °C/30 sec, hybridization 55 °C/30 sec, extension 72 °C/45 sec; final extension 72 °C/2 min. The reference strain JM 109 of Escherichia coli (Promega, REF L2004) was used as a positive control. Fragments of amplified DNA (248 bp) to identify E. coli strains were separated by their molecular weight by electrophoresis - agarose 1 %. The fragments were analyzed by agarose gel staining with Sybr Green (Thermo Fisher) and observed on a Labnet U1001 UV transilluminator, 302 nm, Taiwan.
Antimicrobial susceptibility test
For the evaluation of susceptibility/resistance of E. coli strains to antibiotics, the Kirby-Bauer methodology was used, where parameters established for bacteria isolated from animals by the Clinical and Laboratory Standards Institute (CLSI) were taken as a reference14 (Table 1). Before carrying out the susceptibility test, specific Mueller Hinton agar (Merck KGaA 64271, Darmstact Germany) was prepared to determine the sensitivity of clinically important pathogens according to the manufacturer’s instructions, and sterilized in an autoclave (Automatic Digital Autoclave AVDA050 Liters, Biogenics Lab, Peru); then it was poured into Petri dishes of 35 mm diameter / 10 mm high and two or three isolated colonies were seeded with a bacteriological loop, incubated using an incubator (Memmert CO2 ICO50 GmbH + Co. KG) under aerobic conditions for 18 h at 37 °C. Antimicrobial susceptibility of all isolated colonies was determined against neomycin-N 30 μg, tetracycline-TE 30 μg, sulfamethoxazole-trimethoprim-SXT 25 μg and enrofloxacin-ENR 5 μg (Discs - Thermo Scientific™); the sensitivity of the isolated strains was classified as sensitive, intermediate or resistant by measuring the halo of inhibition according to the parameters established by the CLSI14.
Table 1 Interpretation of antibiotic sensitivity tests, by disc diffusion method (diameter of inhibition zone)
| Antibiotic | Disc concentration (μg) |
Sensitive | Intermediate resistance |
Resistant |
|---|---|---|---|---|
| Tetracycline | 30 | ≥19 | 15-18 | ≤14 |
| Sulfamethoprim | 25 | ≥16 | 11-15 | ≤10 |
| Neomycin | 30 | ≥17 | 13-16 | ≤12 |
| Enrofloxacin | 5 | ≥23 | 17-22 | ≤16 |
Statistical analysis
Results were analyzed using the Graph Pad Prism 9.3.1 software (Prism Software, Irvine, CA, USA). The normality of the data was determined by “Kolmogorov-Smirnoff”. The analysis of variance (ANOVA), followed by Tukey’s multiple comparisons analysis for evaluations between antibiotics (parameters related to sensitivity, resistance). The information obtained was considered statistically significant at a P<0.05.
Results
From the total samples, a total of 52 with positive growth of E. coli on MacConkey II Sorbitol agar were selected. The molecular identification of E. coli present in feces of calves with diarrhea, the uidA gene encoding the β-glucoronidase enzyme12,13,14 was amplified. The processing of the PCR products was analyzed by 1 % agarose gel electrophoresis, a simple and effective method to separate, identify and purify DNA fragments with a molecular size of 0.5 to 25 kb. Electrophoretic bands of the expected size were observed: 248 bp positive for the amplified region of the uidA gene. It was detected in the 52 samples analyzed, evidencing the identification of E. coli, since this gene is specific to the bacterium (Figure 2), of which 63.30 % (n= 35) corresponded to male calves and 32.69 % (n= 17) came from female calves.

Lane 1 100 bp marker. Positive samples in all lanes n= 52.
Figure 2 Amplification of the uidA gene from calf fecal samples by (1 %) agarose gel electrophoresis
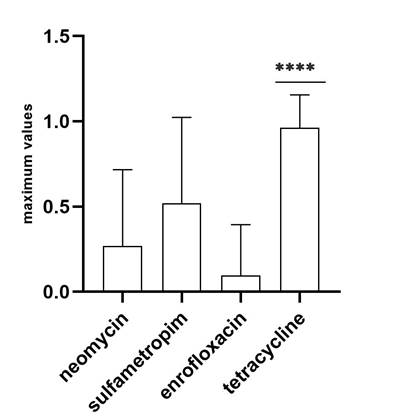
The presentation of susceptibility/resistance to drugs of diarrheal fecal samples from calves was analyzed, where different percentages of resistance could be observed; most strains of E. coli showed resistance to tetracycline (96.15 %, 50/52), likewise, more than 50 % of the samples were resistant to sulfamethoxazole-trimethoprim (51.92 %, 27/52), followed by a significant percentage of resistance to neomycin (26.92 %, 14/52), in addition, less resistance to enrofloxacin was observed (9.61 %, 5/52) (Figure 3).
The presentation of antibiotic resistance was also determined in terms of the variation of the strains isolated in each calf, most had resistance to one (42.30 %, 22/52) and two (30.76 %, 16/52) drugs, the problem worsens in a significant number of calves, presenting multiple resistance in the isolated strains of E. coli, observing resistance to three (19.23 %, 10/52) and four (5.76 %, 3/52) drugs, with the resistance to tetracycline being the most common in all; in addition, resistance to enrofloxacin was the one that occurred in the lowest proportion in the isolated strains (Figure 4).
It was observed that both males and females presented resistance, with higher percentages of resistance observed in males for tetracycline (68 %) and neomycin (64.28 %) respectively; but curiously, regarding females, it could be observed that resistance to the drugs sulfamethoprim (77.70 %), enrofloxacin (80 %) was higher compared to males (Figure 5).
The statistical analysis revealed a significant difference (P<0.0001) regarding the presentation of resistance of E. coli strains from samples of calves with diarrhea to tetracycline (Figure 6).
Discussion
E. coli is one of the main bacterial agents that cause urinary infections in animals, septicemia and diarrhea in farm animals; the phenomenon of resistance expressed by E. coli strains to the drugs used in their control shows therapeutic failure, in addition, many cases of multiple resistance are being observed, which increases worldwide, with the dissemination of resistance becoming a public health problem15,16.
In this research, it was possible to determine different characteristics of resistance to antibiotics and with different percentages, which has allowed determining that the therapeutic failure to antibiotics expressed by the bacterium in calves raised in dairy cattle farms in the region of Cajamarca, Peru, is widespread, being able to identify that all isolated strains of E. coli show resistance; thus, it was possible to observe a higher percentage of resistance to tetracycline and with a significant percentage of multiple resistance with a higher prevalence to two drugs, followed by three and four antibiotics evaluated.
The data obtained on antibiotic resistance in the Cajamarca region allows to mention that it is due to the lack of sanitary control records in the herds, making it difficult to trace the drugs used and, according to the version of the owners, some of these animals when presenting the signs of the disease are treated with antibiotics and others are not, however, all presented resistance to at least one of the antibiotics analyzed, which allows to mention that resistance could be due to the misuse of antibiotics by farmers, being used frequently17, and it is advisable to monitor the calf by electrolyte reconstitution and, if possible, not to administer antibiotics due to the presentation of resistance to these drugs18.
Another possible cause of this widespread resistance to antibiotics could be that the majority of treated calves and mainly in untreated calves, possibly this phenomenon is due to the fact that milk and colostrum from cows that have been treated with an antibiotic facilitates the presence of antibiotic residues in milk, increasing the selection pressure of E. coli strains, with which resistant strains are selected, a practice well established in the regional livestock farming of Cajamarca19,20,21, in addition, it can be assumed that there is diffusion of genes between commensal and pathogenic strains of antibiotic resistance among animals and herds, increasing resistance levels22,23,24.
The pattern of resistance observed in isolated strains of E. coli has an order from highest to lowest prevalence tetracycline, sulfamethoprim, neomycin and enrofloxacin in a smaller proportion, common results that were also obtained by other researchers who report a pattern of resistance to tetracyclines, sulfonamides, penicillins and fluoroquinolones25, cephalothin, tetracycline, trimethoprim-sulfadiazine, ampicillin26, the most common multiresistance pattern was ampicillin-kanamycin-streptomycin-sulfamethoxazole-tetracycline27.
The most frequent resistance phenotype of E. coli strains was to tetracycline 96.15 % (50/52), this is possibly due to the fact that E. coli strains carry tetracycline-resistant phenotypes due to the inappropriate use of the drug by producers, which has generated greater selection pressure in strains carrying genes that confer resistance to tetracycline, contributing to the transfer of antimicrobial resistance genes through strain diffusion28; in addition, these strains are possible sources that spread resistance to the environment when manure is spread in grazing areas as fertilizer29.
The presence of resistance of local strains of E. coli to tetracycline is widespread based on its broad-spectrum use in animal health, as a reservoir of gram-negative bacteria with genes of resistance to tetracycline as a source of infection and with a higher prevalence in E. coli causing diarrhea in calves, a frequent problem that has also been reported in other studies with similar results30,31,32.
It is very important to determine the genes that are involved in these resistance processes related to the use of these drugs, such as adhesion, iron transporters genes33,34, as is the case of the iucD gene9,35, in addition, the processes of horizontal gene transfer, selection pressure, caused by the indiscriminate use of antibiotics36,37, all of the above is necessary knowledge to evaluate the processes of treatment and control of diarrhea in calves in the region of Cajamarca.
Conclusions and implications
The strains of E. coli causing neonatal diarrhea in calves in the Cajamarca region present a prevalence of multiple resistance to the drugs used by farmers in their control, observing a profile of resistance to tetracycline, sulfamethoprim, neomycin and enrofloxacin (TSNE), as a result of the misuse of drugs which increases the selection pressure on strains that promote the expression of genes of virulence and resistance to antibiotics, becoming foci of transmission of resistance to both animals and humans due to the possibility of horizontal transmission between microorganisms. Considering that a definitive diagnosis should be made, determining the etiological agent and susceptibility to antibiotics, then correctly apply the selected antibiotic in correct dose and frequency. In addition, based on the results obtained, it is necessary to determine the resistance genes involved in multiple resistance to antibiotics.
Literatura citada
1. Foster DM, Smith GW. Pathophysiology of diarrhea in calves. Vet Clin North Am Food Anim Pract 2009;(25):13-36. [ Links ]
2. Blanchard PC. Diagnostics of dairy and beef cattle diarrhea. Vet Clin North Am Food Anim Pract 2012;(28):443-464. https://doi.org/10.1016/J.CVFA.2012.07.002. [ Links ]
3. Kolenda R, Burdukiewicz M, Schierack, P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front Cell Infect Microbiol 2015;(5):1-12. https://doi.org/10.3389/fcimb.2015.00023. [ Links ]
4. Barlow J. Mastitis therapy and antimicrobial susceptibility: A multispecies review with a focus on antibiotic treatment of mastitis in dairy cattle. J Mammary Gland Biol Neoplasia 2011;(6):383-407. https://doi.org/10.1007/s10911-011-9235-z. [ Links ]
5. Mcewen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis 2002;(34):3:S93-S106. [ Links ]
6. Ma F, Xu S, Tang Z, Li Z, Zhang L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosafety and Health 2021; (3):32-38. https://doi.org/10.1016/J.BSHEAL.2020.09.004. [ Links ]
7. Jarrige N, Cazeau G, Bosquet G, Bastien J, Benoit F, Gay E. Effects of antimicrobial exposure on the antimicrobial resistance of Escherichia coli in the digestive flora of dairy calves. Prev Vet Med 2020;(185):105177. https://doi.org/10.1016/j.prevetmed.2020.105177. [ Links ]
8. Gharieb R, Fawzi E, Elsohaby I. Antibiogram, virulotyping and genetic diversity of Escherichia coli and Salmonella serovars isolated from diarrheic calves and calf handlers. Com Immunol Microbiol Infect Dis 2019;(67):101367. https://doi.org/10.1016/j.cimid.2019.101367. [ Links ]
9. Louge-Uriarte EL, González-Pasayo RA, Massó M, Carrera-Paez L, Domínguez-Moncla M, Donis N, et al. Molecular characterization of multidrug-resistant Escherichia coli of the phylogroups A and C in dairy calves with meningitis and septicemia. Microbial Pathogenesis 2022;(163):105378. https://doi.org/10.1016/J.MICPATH.2021.105378. [ Links ]
10. Resapath A. French surveillance network for antimicrobial resistance in pathogenic bacteria of animal origin. ANSES. 2018. [ Links ]
11. Instituto de Estándares Clínicos y de Laboratorio (CLSI). Performance standards for antimicrobial susceptibility testing: Sixteenth Informational Suppl. Wayne, PA: 2006. [ Links ]
12. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000;181(1):261-72. https://doi.org/dqjh6j. [ Links ]
13. Konno, T; Yatsuyanagi, J; Takahashi, S; Kumagai, Y. Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Jpn J Infect Dis 2012;65:203-207. https://doi.org/f3zvj5. [ Links ]
14. Instituto de Estándares Clínicos y de Laboratorio (CLSI). The User's Guide for EUCAST and CLSI-potency Neo-Sensitabs™. http://pishrotashkhis.com/wp-content/uploads/2017/07/Neo-SENSITAB-CLSI-EUCAST-Potency.pdf. [ Links ]
15. Allocati N, Masulli M, Alexeyev MF, Di Ilio C. Escherichia coli in Europe: an overview. Int J Environ Res Public Health 2013;25(12):6235-54. doi: 10.3390/ijerph10126235. PMID: 24287850; PMCID: PMC3881111. [ Links ]
16. Astorga F, Navarrete-Talloni MJ, Miró MP, Bravo V, Toro M, Blondel CJ, Hervé-Claude LP. Antimicrobial resistance in E. coli isolated from dairy calves and bedding material. Heliyon 2019;26:(11):e02773. doi: 10.1016/j.heliyon.2019.e02773. PMID: 31844709; PMCID: PMC6888714. [ Links ]
17. Berge AC, Moore DA, Sischo WM. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl Environ Microbiol 2006;72(6):3872-8. doi: 10.1128/AEM.02239-05. PMID: 16751491; PMCID: PMC1489621. [ Links ]
18. Constable PD. Antimicrobial use in the treatment of calf diarrhea. J Vet Intern Med 2004;18(1):8-17. doi:10.1892/0891-6640(2004)18<8:auitto>2.0.co;2. [ Links ]
19. Duse A, Waller KP, Emanuelson U, Unnerstad HE, Persson Y, Bengtsson B. Farming practices in Sweden related to feeding milk and colostrum from cows treated with antimicrobials to dairy calves. Acta Vet Scand 2013;9:55(1):49. doi:10.1186/1751-0147-55-49. PMID: 23837498; PMCID: PMC3720286. [ Links ]
20. Brunton LA, Duncan D, Coldham NG, Snow LC, Jones JR. A survey of antimicrobial usage on dairy farms and waste milk feeding practices in England and Wales. Vet Rec 2012;22:171(12):296. doi:10.1136/vr.100924. [ Links ]
21. Duse A, Waller KP, Emanuelson U, Unnerstad HE, Persson Y, Bengtsson B. Risk factors for antimicrobial resistance in fecal Escherichia coli from preweaned dairy calves. J Dairy Sci 2015;98(1):500-16. doi:10.3168/jds.2014-8432. [ Links ]
22. Sørum H, Sunde M. Resistance to antibiotics in the normal flora of animals. Vet Res 2001;32(3-4):227-41. doi:10.1051/vetres:2001121. [ Links ]
23. Sunde M, Fossum K, Solberg A, Sørum H. Antibiotic resistance in Escherichia coli of the normal intestinal flora of swine. Microb Drug Resist 1998;4(4):289-99. doi: 10.1089/mdr.1998.4.289. [ Links ]
24. Liu J, Yu F, Call DR, Mills DA, Zhang A, Zhao Z. On-farm soil resistome is modified after treating dairy calves with the antibiotic florfenicol. Sci Total Environ 2015;750:141694. doi: 10.1016/j.scitotenv.2020.141694. [ Links ]
25. Ferroni L, Albini E, Lovito C, Blasi F, Maresca C, Massacci FR, et al. Antibiotic consumption is a major driver of antibiotic resistance in calves raised on Italian cow-calf beef farms. Res Vet Sci 2022;145:71-81. doi:10.1016/j.rvsc.2022.01.010. [ Links ]
26. Rigobelo EC, Gamez HJ, Marin JM, Macedo C, Ambrosin JA, Ávila FA. Fatores de virulência de Escherichia coli isolada de bezerros com diarréia Vet Medicine • Arq. Bras Med Vet Zootec 2006;58(3) https://doi.org/10.1590/S0102-09352006000300003. [ Links ]
27. Gow SP, Waldner CL, Rajić A, McFall ME, Reid-Smith R. Prevalence of antimicrobial resistance in fecal generic Escherichia coli isolated in western Canadian cow-calf herds. Part I-beef calves. Can J Vet Res 2008;72(2):82-90. [ Links ]
28. Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, et al. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol 2002;68(2):576-81. doi: 10.1128/AEM.68.2.576-581.2002. [ Links ]
29. Barour D, Berghiche A, Boulebda N. Antimicrobial resistance of Escherichia coli isolates from cattle in Eastern Algeria. Vet World 2019;12(8):1195-1203. doi: 10.14202/vetworld.2019.1195-1203. [ Links ]
30. Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, Wagner DD, McDermott PF, Walker RD, Meng J. Resistencia antimicrobiana de Escherichia coli O157 aislada de humanos, bovinos, porcinos y alimentos. Appl Environ Microbiol 2002;68(2):576-81. doi:10.1128/AEM.68.2.576-581.2002. [ Links ]
31. Barour D, Berghiche A, Boulebda N. Antimicrobial resistance of Escherichia coli isolates from cattle in Eastern Algeria. Vet World 2019;12(8):1195-1203. doi: 10.14202/vetworld.2019.1195-1203. [ Links ]
32. Formenti N, Martinelli C, Vitale N, Giovannini S, Salogni C, Tonni M, et al. Antimicrobial resistance of Escherichia coli in dairy calves: a 15-year retrospective analysis and comparison of treated and untreated animals. Animals (Basel) 2021;11(8):2328. doi:10.3390/ani11082328. [ Links ]
33. Watts RE, Totsika M, Challinor VL, Mabbett AN, Ulett GC, Voss JJ. De Schembri MA . Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infection and Immunity 2012;80(1): 333-344. https://doi.org/10.1128/IAI.05594-11. [ Links ]
34. Su Q, Guan T, Lv H. Siderophore biosynthesis coordinately modulated the virulence-associated interactive metabolome of uropathogenic Escherichia coli and human urine. Scientific Reports 2016;6(1):24099. https://doi.org/10.1038/srep24099. [ Links ]
35. Subashchandrabose S, Mobley H LT. Host -Pathogen Interface 80 during urinary tract infection. Metallomics 2015;7(6):935-942. https://doi.org/10.1039/C4MT00329B. [ Links ]
36. Jia Y, Mao W, Liu B, Zhang S, Cao J, Xu X. Study on the drug resistance and pathogenicity of Escherichia coli isolated from calf diarrhea and the distribution of virulence genes and antimicrobial resistance genes. Front Microbiol 2022;3:992111. doi:10.3389/fmicb.2022.992111. [ Links ]
37. Shin SW, Shin MK, Jung M, Belaynehe KM, Yoo HS. Prevalence of antimicrobial resistance and transfer of tetracycline resistance genes in Escherichia coli isolates from beef cattle. Appl Environ Microbiol 2015;81(16):5560-6. doi: 10.1128/AEM.01511-15. [ Links ]
Received: November 17, 2022; Accepted: May 05, 2023











 texto en
texto en