Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ciencias pecuarias
versão On-line ISSN 2448-6698versão impressa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.14 no.2 Mérida Abr./Jun. 2023 Epub 26-Jun-2023
https://doi.org/10.22319/rmcp.v14i2.6315
Technical notes
Genetic analysis of Oaxacan Mixteco Creole cattle
a Universidad Autónoma Benito Juárez de Oaxaca. Facultad de Medicina Veterinaria y Zootecnia. Cuerpo Académico Ciencias Veterinarias Aplicadas al Desarrollo Regional. Av. Universidad S/N. Ex-Hacienda 5 Señores, 68120. Oaxaca de Juárez, Oaxaca, México.
b Universidad Tecnológica de la Mixteca, Brigada de Promoción del Desarrollo. Oaxaca, México.
c Universidad de Córdoba, Facultad de Veterinaria. Córdoba, España.
The Mixteco Creole cattle is a little explored genetic resource, which, however, has great value due to its potential to be used in production systems that are respectful of the environment and adaptable to its conditions. The identification and characterization of this local resource is essential for its conservation and improvement. For this reason, in the present study it was carried out the analysis of the diversity and genetic relationships of the Mixteco Creole cattle population of Oaxaca, using 19 microsatellite DNA markers and 32 reference cattle populations belonging to the BIOBOVIS consortium of the CONBIAND Network. The mean number of alleles detected was 8.8 ± 2.1 and the estimated effective number of alleles was 4.5 ± 1.2. The genetic diversity represented by the expected (0.7700 ± 0.0682) and observed (0.7170 ± 0.0998) heterozygosity values was within the range of estimators obtained in previous studies with local cattle populations using microsatellite markers. An analysis of the population structure revealed a predominant influence of Iberian germplasm (Bos taurus). There is also a close relationship between the Mixteco Creole and the rest of the Mexican Creole cattle populations, with the exception of the Tropical Dairy Creole.
Keywords Conservation; Creole cattle; Genetic characterization; Microsatellites
El bovino Criollo Mixteco es un recurso genético poco explorado, pero de gran valor por su potencial para ser empleado en sistemas de producción respetables con el entorno y adaptables a las condiciones del mismo. La identificación y caracterización de este recurso local es un punto primordial para su conservación y mejora, es por esto que en el presente estudio se llevó a cabo el análisis de la diversidad y relaciones genéticas de la población de bovinos Criollos Mixtecos de Oaxaca, mediante el empleo de 19 marcadores de ADN microsatélites y 32 poblaciones bovinas de referencia, pertenecientes al consorcio BIOBOVIS de la Red CONBIAND. El número medio de alelos detectados fue de 8.8 ± 2.1 y el número efectivo de alelos estimado fue de 4.5 ± 1.2. La diversidad genética representada por los valores de heterocigosidad esperada (0.7700 ± 0.0682) y observada (0.7170 ± 0.0998), se encontró dentro del rango de estimadores obtenidos en estudios previos con poblaciones bovinas locales empleando marcadores microsatélites. El análisis de la estructura poblacional reveló una influencia predominante de germoplasma ibérico (Bos taurus). Se observa además una estrecha relación entre el Criollo Mixteco y el resto de poblaciones bovinas criollas mexicanas a excepción del Criollo Lechero Tropical.
Palabras clave Caracterización genética; Microsatélites; Conservación; Bovino criollo
Creole cattle represent a genetic resource of great importance for the supply of food and raw materials in areas with extreme climatic conditions, scarce food resources and high incidence of infectious and parasitic diseases1, and potentially contribute to hunger and poverty reduction, as well as to sustainable development2. However, the inability to appreciate the real biological, economic and cultural value of these animals has caused an aggressive extension of highly selected breeds, causing a constant erosion that endangers the existence of these resources and, thereby, an irreparable loss of genetic variability, which could be of great value to face the effects of climate change.
In the state of Oaxaca, there is a population of Creole cattle located in the Mixteca region (Figure 1A), known as Mixteco Creole. Phenotypically, they are medium-sized animals, with an average height at the withers of 1.03 ± 0.16 m and an average weight of 176 ± 51.48 kg (parameters reported for an age of 1 to 3 yr)3; their coat can be uniform black or red, or black- or red-spotted (Figure 1B). The origin of the Mixteco Creole cattle dates back to colonial times, having probably been brought by the first Spaniards for the construction of the convents along the Oaxacan territory, on what is currently known as the Dominican Route4.
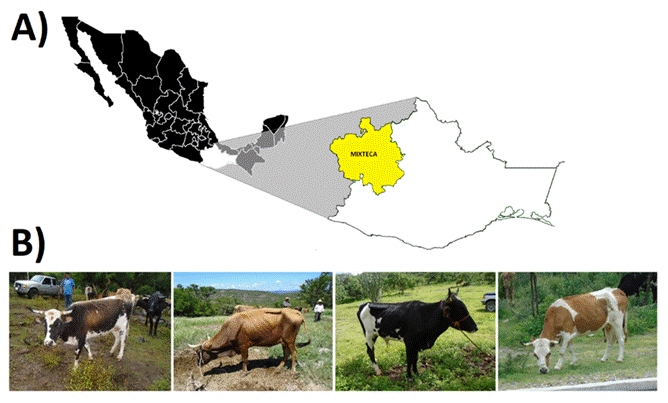
The image shows the coloration and morphostructural characteristics of the Mixteco Creole bovine cattle
Figure 1 A) Geographic location of the Mixteca region of Oaxaca. B) Mixteco Creole bovine cattle
Since its introduction into the region, the Mixteco Creole cattle have successfully adapted to the geographical and environmental conditions prevailing in the area, which is characterized by a complicated orography and fluctuations in the availability of food and water. However, these cattle are capable of being productive under such conditions, which makes them suitable for developing environmentally friendly, resilient production systems that are able to cope with changes in the environment, especially with the current trend in Latin America towards the development of more intensive and sustainable production systems5.
According to the Food and Agriculture Organization of the United Nations (FAO), the identification and genetic characterization of local livestock is an essential step in the conservation and use of these genetic resources6. In this regard, microsatellite molecular markers have proven to be a highly effective tool to genetically characterize Creole cattle populations in the Americas7-10, Therefore, this study evaluated the genetic diversity of the Mixteco Creole cattle population and their genetic relationships with other local and specialized cattle populations, using 19 microsatellite markers and 32 reference populations in order to generate information on the conservation status of this valuable local genetic resource.
A total of 40 adult Mixteco Creole cattle (29 females and 11 males) were selected and identified based on phenotypic characteristics of Creole cattle previously reported in the literature, including color pattern, size, and zoometric parameters3,11; those individuals that exhibited phenotypic characteristics typical of Zebu breeds were discarded. In order to avoid close kinship relationships among the selected individuals, only one specimen was included for each one of the cattle production units sampled, geographically separated in different communities of the Oaxacan Mixteca (17°48′00″ N, 97°46′00″ W), in addition to confirming, through an interview, the absence of genetic connections (use of stallions) between the production units. The sample size was defined taking as a reference the information published for population genetic studies using microsatellite molecular markers12, as well as the sample size suggested by FAO for genetic characterization studies of local livestock populations using microsatellites (n=25 to 40)13.
The biological material consisted of whole blood samples with anticoagulant (EDTA) obtained by aseptic puncture of the jugular vein, from which nucleic acids were extracted using the ReliaPrepTM Blood gDNA Miniprep System kit (Promega), following the manufacturer's instructions. A panel of 19 microsatellite markers recommended by FAO-ISAG was used for the genetic analysis of the population14 (Table 1), having been amplified by PCR and processed in an ABI377XL capillary sequencer (Applied Biosystems), with subsequent allelic typing, following the methodology established at the Laboratory for the Improvement and Conservation of Animal Genetic Resources of the University of Cordoba, Spain7.
Table 1 Allele frequencies observed in the Mixteco Creole Cattle population
| BM1818 | BM1824 | BM2113 | CSRM60 | CSSM66 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | Frec. | Allele | Frec. | Allele | Allele | Frec. | Allele | Frec. | Allele |
| 260 | 0.0132 | 179 | 0.2750 | 126 | 0.0385 | 91 | 0.0125 | 179 | 0.0132 |
| 262 | 0.0395 | 181 | 0.1750 | 128 | 0.0513 | 93 | 0.1500 | 181 | 0.0921 |
| 264 | 0.3553 | 183 | 0.3875 | 130 | 0.0769 | 95 | 0.0125 | 183 | 0.1974 |
| 266 | 0.1842 | 185 | 0.0125 | 132 | 0.0256 | 97 | 0.1375 | 185 | 0.0789 |
| 268 | 0.3421 | 189 | 0.1500 | 134 | 0.0769 | 99 | 0.0250 | 187 | 0.0132 |
| 270 | 0.0526 | 136 | 0.2436 | 101 | 0.0125 | 189 | 0.2895 | ||
| 272 | 0.0132 | 138 | 0.3077 | 103 | 0.4125 | 191 | 0.0132 | ||
| 140 | 0.1026 | 105 | 0.1875 | 193 | 0.1842 | ||||
| 142 | 0.0769 | 111 | 0.0375 | 197 | 0.1184 | ||||
| 113 | 0.0125 | ||||||||
| ETH003 | ETH010 | ETH185 | ETH225 | HAUT27 | |||||
| Allele | Frec. | Allele | Frec. | Allele | Allele | Frec. | Allele | Frec. | Allele |
| 103 | 0.0263 | 209 | 0.1667 | 220 | 0.0286 | 139 | 0.2000 | 128 | 0.0135 |
| 109 | 0.1053 | 211 | 0.0128 | 222 | 0.0571 | 141 | 0.0125 | 140 | 0.0270 |
| 115 | 0.0658 | 213 | 0.1026 | 226 | 0.0571 | 143 | 0.1125 | 142 | 0.0811 |
| 117 | 0.3421 | 215 | 0.1154 | 228 | 0.6000 | 145 | 0.0250 | 146 | 0.0135 |
| 119 | 0.1974 | 217 | 0.3333 | 230 | 0.0143 | 147 | 0.2625 | 148 | 0.5405 |
| 123 | 0.0658 | 219 | 0.1923 | 232 | 0.1000 | 149 | 0.2875 | 150 | 0.2297 |
| 125 | 0.1711 | 221 | 0.0256 | 234 | 0.1143 | 151 | 0.0125 | 152 | 0.0676 |
| 129 | 0.0263 | 223 | 0.0513 | 236 | 0.0143 | 153 | 0.0250 | 154 | 0.0270 |
| 240 | 0.0143 | 157 | 0.0625 | ||||||
| HEL009 | ILSTS006 | INRA32 | INRA63 | MM12 | |||||
| Allele | Frec. | Allele | Frec. | Allele | Allele | Frec. | Allele | Frec. | Allele |
| 149 | 0.0128 | 287 | 0.0556 | 168 | 0.0152 | 175 | 0.4250 | 105 | 0.0132 |
| 151 | 0.0128 | 289 | 0.0417 | 174 | 0.0152 | 177 | 0.3250 | 109 | 0.0132 |
| 153 | 0.3077 | 291 | 0.2361 | 176 | 0.1364 | 179 | 0.0125 | 117 | 0.1579 |
| 155 | 0.0641 | 293 | 0.2778 | 178 | 0.2879 | 183 | 0.2000 | 119 | 0.1184 |
| 157 | 0.0256 | 295 | 0.1111 | 180 | 0.3485 | 185 | 0.0375 | 121 | 0.2368 |
| 159 | 0.0256 | 297 | 0.2639 | 182 | 0.0303 | 123 | 0.1184 | ||
| 161 | 0.2308 | 299 | 0.0139 | 184 | 0.1212 | 125 | 0.0526 | ||
| 163 | 0.1154 | 186 | 0.0152 | 129 | 0.0132 | ||||
| 165 | 0.0513 | 188 | 0.0303 | 131 | 0.0263 | ||||
| 167 | 0.0513 | 133 | 0.1974 | ||||||
| 169 | 0.0128 | 135 | 0.0395 | ||||||
| 171 | 0.0897 | 139 | 0.0132 | ||||||
| SPS115 | TGLA053 | TGLA122 | TGLA227 | ||||||
| Allele | Frec. | Allele | Frec. | Allele | Frec. | Allele | Frec. | ||
| 242 | 0.0375 | 151 | 0.0658 | 134 | 0.0250 | 79 | 0.0769 | ||
| 244 | 0.4875 | 153 | 0.0395 | 140 | 0.0125 | 81 | 0.0256 | ||
| 246 | 0.0625 | 157 | 0.2105 | 142 | 0.0375 | 83 | 0.1282 | ||
| 248 | 0.1875 | 159 | 0.1711 | 144 | 0.0375 | 85 | 0.3077 | ||
| 250 | 0.0250 | 163 | 0.0526 | 146 | 0.0750 | 87 | 0.0128 | ||
| 252 | 0.0875 | 165 | 0.0921 | 148 | 0.0125 | 89 | 0.0641 | ||
| 254 | 0.0250 | 167 | 0.1842 | 150 | 0.3875 | 91 | 0.0641 | ||
| 256 | 0.0875 | 169 | 0.1447 | 152 | 0.2875 | 93 | 0.0128 | ||
| 175 | 0.0263 | 154 | 0.0125 | 95 | 0.0385 | ||||
| 181 | 0.0132 | 156 | 0.0125 | 99 | 0.2692 | ||||
| 160 | 0.0625 | ||||||||
| 168 | 0.0125 | ||||||||
| 174 | 0.0250 | ||||||||
The information generated from allelic typing was used to calculate allele and genotypic frequencies for each microsatellite marker using the MSTools add-in for Excel (Genetics Dept, TCD, Ireland). The number of alleles per locus, effective number of alleles, observed and expected heterozygosity, polymorphic information content (PIC), and FIS inbreeding coefficient were estimated with Popgene v 1.3215. The Hardy-Weinberg equilibrium test was carried out with Arlequin v 3.116.
The analysis of the population structure and genetic relationships was carried out using allelic information from 32 reference populations previously reported in the literature (Figure 2)17, belonging to the BIOBOVIS Consortium (https://BIOBOVIS.jimdofree.com/) of the Network for the Conservation of Local Domestic Animal Biodiversity (CONBIAND). The reference populations were divided into four groups according to their origin or specialization (Mexican Creoles, Iberians, specialized Europeans, and Zebu). The genetic distance between pairs of NEI populations (DST) was estimated with the Arlequin v3.1 software. Based on the information generated in the genetic distance matrix, a graphical representation in the form of a phylogenetic tree was created using the SplitsTree v.4.14.16 software, with the Neighbor-Joining method. Finally, the Structure software was utilized18 to infer the population structure, using the following parameters: 100,000 warm-up iterations followed by 1'000,000 Markov chain-based Monte Carlo (MCMC) iterations. A total of 32 different runs (K2 to K33) were performed to estimate the most probable number of existing clusters. The optimal K value was estimated by the modal value method of the Delta K distribution, using the formula Delta K = mean (|L''(K)|) / sd (L(K)).
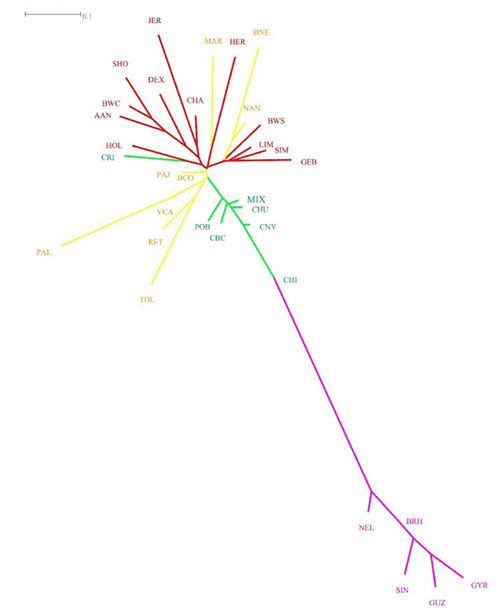
The circle shows the location of the Creole populations within the genetic distance tree (except for the Tropical Dairy Creole), revealing their intermediate position between the European Bos taurus breeds and the Bos indicus breeds. Populations: MIX (Mixteco Creole Bovine), CRI (Tropical Dairy Creole), POB (Poblano Creole), CBC (Baja California Creole), CHU (Chihuahua Creole), CNY (Nayarit Creole), CHI (Chiapas Creole), TDL (Fighting), RET (Very dark), BCO (Red-spotted), BNE (Black-spotted), MAR (Marshland), PAJ (Straw-colored), NAN (Andalusian Black), VCA (Canary Island cattle), PAL (Palmera), AAN (Aberdeen Angus), BWC (British White Cattle), HER (Hereford), JER (Jersey), SHO (Shorthorn), DEX (Dexter), BWS (Brown Swiss), CHA (Charolais), HOL (Holstein- Fresian), LIM (Limousin), SIM (Simmental), GEB (Gelbvieh), GYR (Gyr), BRH (Brahman), SIN (Sindi), GUZ (Guzerat), NEL (Nelore).
Figure 2 NEI's genetic distance (unbiased distance) tree using the Neighbor-Joining method
Table 1 shows the results of the calculation of allele frequencies for each of the 19 loci analyzed in the Mixteco Creole cattle population. Polymorphic variation was observed in all the loci analyzed. In total, 168 alleles were detected distributed among the 19 microsatellites, representing a mean number of alleles of 8.8 ± 2.1, with TGLA122 (Na= 13), MM12 (Na= 12) and HEL009 (Na= 12) markers having the highest number of alleles (Table 2). With regard to the effective number of alleles, a mean of 4.5 ± 1.2 alleles was observed, while the average polymorphic information content was 0.7286 ± 0.0748. The observed heterozygosity value was 0.7170 ± 0.0998, and the expected heterozygosity was 0.7700 ± 0.0682. Table 2 shows the results of the main genetic diversity parameters for each of the microsatellite markers evaluated. The HAUT27, ILSTS006, and TGLA227 markers were the only loci in Hardy Weinberg disequilibrium (P<0.05). On the other hand, 15 of the 19 microsatellite markers showed positive FIS values, and the remaining 4 showed negative values; however, most of them were separated from the zero value, obtaining a mean FIS value of 0.058, with TGLA227 and BM1818 markers showing the greatest deviation from the positive FIS value and CSRM60 marker showing the greatest deviation from the negative FIS value.
Table 2 Genetic diversity parameters
| Marker | Na | Ne | PIC | Ho | He | FIS | P value |
|---|---|---|---|---|---|---|---|
| BM1818 | 7 | 3.5479 | 0.6695 | 0.5789 | 0.7277 | 0.1938 | 0.0640 |
| BM1824 | 5 | 3.5834 | 0.6728 | 0.6500 | 0.7301 | 0.0984 | 0.7939 |
| BM2113 | 9 | 5.3462 | 0.7907 | 0.7949 | 0.8235 | 0.0222 | 0.8494 |
| CSRM60 | 10 | 4.0100 | 0.7195 | 0.8250 | 0.7601 | -0.0991 | 0.6078 |
| CSSM66 | 9 | 5.3780 | 0.7895 | 0.8684 | 0.8249 | -0.0668 | 0.4087 |
| ETH003 | 8 | 4.8456 | 0.7673 | 0.7632 | 0.8042 | 0.0384 | 0.8776 |
| ETH010 | 8 | 4.9223 | 0.7704 | 0.7436 | 0.8072 | 0.0668 | 0.1559 |
| ETH185 | 9 | 2.5574 | 0.5860 | 0.5143 | 0.6178 | 0.1555 | 0.2228 |
| ETH225 | 9 | 4.7690 | 0.7597 | 0.7750 | 0.8003 | 0.0194 | 0.9550 |
| HAUT27* | 8 | 2.7939 | 0.6022 | 0.6216 | 0.6509 | 0.0319 | 0.0042 |
| HEL009 | 12 | 5.5410 | 0.7990 | 0.8205 | 0.8302 | -0.0012 | 0.8609 |
| ILSTS006* | 7 | 4.5474 | 0.7458 | 0.8056 | 0.7911 | -0.0326 | 0.0136 |
| INRA32 | 9 | 4.1644 | 0.7244 | 0.6667 | 0.7716 | 0.1227 | 0.4309 |
| INRA63 | 5 | 3.0505 | 0.6101 | 0.6000 | 0.6807 | 0.1074 | 0.4384 |
| MM12 | 12 | 6.5045 | 0.8283 | 0.8421 | 0.8575 | 0.0049 | 0.2746 |
| SPS115 | 8 | 3.3934 | 0.6763 | 0.6750 | 0.7142 | 0.0430 | 0.7280 |
| TGLA053 | 10 | 6.8274 | 0.8366 | 0.7368 | 0.8649 | 0.1367 | 0.1248 |
| TGLA122 | 13 | 4.0455 | 0.7211 | 0.7000 | 0.7623 | 0.0702 | 0.5162 |
| TGLA227* | 10 | 4.9951 | 0.7743 | 0.6410 | 0.8102 | 0.1985 | 0.0017 |
Na= number of alleles; Ne= effective number of alleles; PIC= polymorphic information content; Ho= observed heterozygosity; He= expected heterozygosity; FIS= coefficient of inbreeding; P value = Hardy Weinberg equilibrium significance.
The genetic distance analysis revealed that the Mixteco Creole cattle clusters with Mexican Creole populations (Figure 2), showing a smaller distance with regard to Creole cattle from Chihuahua (D= 0.046), Baja California (D= 0.064), and Puebla (D= 0.078). As for the rest of the population groups, the Mixteco Creole exhibited a smaller genetic distance in relation to the Red-spotted population (D= 0.103), belonging to the Spanish landrace group, as well as with respect to the Limousin breed (D= 0.190), which is one of the specialized European breeds. The greatest genetic distance was observed with each of the Bos indicus breeds (Nelore 0.588 - Gyr 0.734).
Finally, the results of the analysis of the population structure through the assignment test with Structure software (Figure 3A) and the subsequent calculation of the optimal K value using the modal value method of the Delta K distribution (Figure 3B), revealed that the optimal K was 8. The proportions of assignment to each cluster for Mixteco Creole are shown in Figure 3A. According to the results of K8, it is observed that, with the exception of the Dairy Creole population, the rest of the Mexican Creole populations, including the Mixteco Creole, exhibit a higher percentage of assignment to cluster 1, which includes the local Black-spotted, Red-spotted, Andalusian Black, and Straw-colored Spanish populations (Figure 3). The rest of the Mixteco Creole cattle genome was distributed as follows: 14.2 % with the rest of Spanish landraces, 13.8 % with specialized European breeds, and 9.9 % with the cluster of Zebu populations.
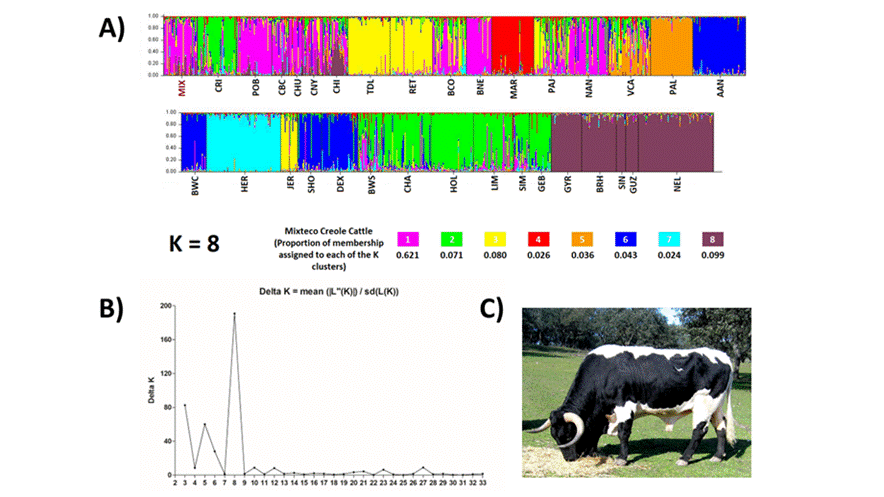
(Source of the photograph: National group of Black-spotted and Red-spotted cattle breeders’ associations).
Figure 3 A) Results of the Bayesian Structure analysis for the Mixteco Creole Cattle population, using 32 reference populations and a total of 8 inferred clusters (K8). B) Estimation of the optimal K using the modal value method of the Delta K distribution. C) Individual of the Black-spotted breed
Local populations play a key role in livestock breeding, as they have been the basis for the development of specialized breeds and today constitute a reservoir of genetic diversity that must be preserved19. In this sense, the Mixteco Creole cattle represents a local livestock resource of great value that must be preserved, for which purpose its genetic evaluation is essential.
The results of allelic diversity, represented by the mean number of alleles observed, showed a similarity with the data reported in previous studies in bovine populations, when compared with the mean values per population [6.92 ± 0.99]7, [6.78 ± 1.88]8, [8.31 ± 2.10]20, however, they are lower when compared with the mean number of alleles detected considering the Creole populations as a group [14.21 ± 3.74]8, [15.5 ± 0.9]17. The difference observed in the mean number of alleles when compared at the group level is due to the heterogeneity of Creole populations in the Americas, which has been confirmed in several studies using autosomal and mitochondrial polymorphisms21-23. This heterogeneity is probably the result of several factors such as differences in the origin of the populations17, differentiation by geographical location18, as well as the process of genetic drift and the contribution of animals of different origins, which have been mixed at some point with the Creole populations, as has been described with the introgression of Zebu, African and British breeds21,23-24. This data should be taken with caution, since the heterogeneity observed could also reflect the state of threat to the population, due to dilution as a result of intensive interbreeding or as a consequence of isolation and abandonment17.
The values of observed and expected heterozygosity represent a measure of genetic diversity in a population; however, they are estimated using different data. On the one hand, expected heterozygosity is estimated based on allele frequencies, while observed heterozygosity is estimated from genotypic frequencies, so the differences observed between both estimates can be an indicator of the level of inbreeding in the population25. The present study showed a difference between heterozygosity values; the observed heterozygosity was lower than the expected heterozygosity, suggesting a tendency towards inbreeding, as could be subsequently verified in the estimation of the FIS value. On the other hand, when comparing the expected heterozygosity values and the effective number of alleles with the values reported for the Creole breeds using a similar panel of microsatellites10,17,20, a correspondence in the results was observed. The effective number of alleles represents the number of alleles expected in a population with the same heterozygosity but with equally distributed allele frequencies26; therefore, if the allele frequencies are highly unbalanced and only some alleles are in the majority, the effective number of alleles will tend to be lower27, as was observed in the Mixtec Creole population, among which certain alleles are more frequent. The relevance of estimating the number of alleles and effective number of alleles lies in the fact that these data can be used as a conservation criterion, since allelic diversity can have important implications in the response to selection for adaptation to changing environments28, This is of great importance when talking about Creole populations, as they are considered reservoirs of genetic information to face potential environmental alterations due to climate change, so it is of great importance to implement measures for the conservation of allelic diversity.
The polymorphic information content exceeded 0.5 for all markers, which, according to the scale proposed by Botstein29, indicates that the microsatellites evaluated are highly informative, and could be used for further genetic diversity monitoring studies or parentage testing. Regarding Wright's fixation index, the estimation of this parameter provides a measure of the degree of inbreeding of individuals with respect to the population to which they belong30. Although generally positive, the FIS value can be negative if inbreeding is systematically avoided within populations31. In the present study, the estimated FIS values were mostly positive, far from zero, suggesting a tendency for heterozygote deficiency inbreeding. The result observed in the population, in which the deviation of FIS with respect to the zero value is positive can be attributed to the condition of the domestic population, where mating is not random and the proportion of males is lower compared to females, in addition to being long production systems with few animals and whose replacements are usually obtained within the same production units, which predisposes to inbreeding32-33. On the other hand, with respect to the Hardy-Weinberg equilibrium test, of the 19 markers analyzed, only markers HAUT27, ILSTS006 and TGLA227 were found to be out of equilibrium; this data increases the reliability of the results obtained in the study and also suggests that the population is not being subjected to perturbing forces that cause significant changes in their genotypic frequencies34.
Genetic relationships between populations were analyzed using genotypic information from 32 breeds belonging to the BIOBOVIS consortium of the CONBIAND Network, which were selected for their potential relationship with Mixteco Creole cattle. We used 6 Mexican Creole populations, 9 Spanish local populations, which could be found among the founding populations of Latin American cattle, 12 European specialized breeds, and 5 Zebu breeds. As Figure 2 shows, the results of the genetic distance calculation show that the Mixteco Creole groups with the Mexican Creole populations of Chihuahua, Nayarit, Baja California and Puebla, separated from the European and Zebu cattle groups. This distribution has been reported in previous work with Latin American Creole cattle17,35 This shows that, like other Creole populations in the Americas, the Mixteco Creole has an identity closely related to other Creole cattle due to their shared origin; this identity has been preserved despite the geographic distance between the populations. Interestingly, the smallest genetic distance to a non-Creole population was observed with the Red-spotted population, which has been described as one of the possible founding populations of American Creole cattle20. Currently, in the Mixtec region, the Zebu breeds have been replaced by Bos taurus cattle of better temperament, as, according to producers, crossbreeding between Zebu and Creole cattle generated individuals with a temperament that was difficult to manage, while European cattle, having been subjected to more intensive selection for docility and ease of management36, do not exhibit this issue. This fact has probably prevented the introgression of Zebu germplasm into the Creole population from increasing, maintaining the genetic distance between these populations.
Regarding the population structure, Bayesian analysis was carried out with Structure software, calculating different values of K (2 - 33), with the subsequent estimation of the optimal K (K= 8). A model with K= 8 populations was suggested, because it was associated with a higher probability (Figure 3B), suggesting that there are closely related races or groups (Figure 3B)7. For a K = 8, the results indicate that Mixteco Creole cattle maintain a relatively uniform population structure, sharing 62 % of their genome with Mexican Creole populations, except for the Tropical Dairy Cattle. Similarly to what was reported for Ecuadorian local breeds, a contribution of Red-spotted and Straw-colored cattle and a low relevance of Marshland cattle were observed20, which suggests that the Mixteco Creole is integrated in its origin with Latin American Creole populations and reinforces the theory that these breeds are part of the founding populations of Creole cattle in Latin America, since, as has been described in other works, most of the Spanish cattle that gave rise to the Creole populations in America came from southern Spain20. In addition, this cluster also includes the Spanish Black-spotted (Figure 3C) and Andalusian Back landraces, which is interesting because the characteristic coloration patterns of the Mixteco Creole are similar to the coloration patterns of these Iberian populations, mainly in the Black-spotted population, whose genome percentage assigned to this cluster was 87.4 %. The Mixteco Creole genome composition for K= 8 exhibited genetic heterogeneity, which has been previously described in other Creole populations using mitochondrial, autosomal and Y chromosome markers22,24,37. The results confirm that the influence of Iberian cattle is predominant in the Mixteco Creole cattle, as is the case in the Creole populations of the Americas, which retain genetic signatures of their Iberian ancestry21. However, it is also possible to infer that there may have been recent contributions of exotic germplasm belonging to cattle of different origins, which, based on the percentages of allocation observed in lower proportions, would be European and Zebu. The presence of Zebu germplasm may be the remnant of the importation of bulls used as sires for females of Creole breeds, a practice of indiscriminate crossbreeding widely used in several Latin American countries since the middle of the last century22,33, but which, as mentioned above, is not currently common among producers in the Mixteca region. Another possible cause for the presence of Zebu germplasm may be related to the flow of ancestral genes between African Zebu cattle and Iberian cattle before they were brought to America, as has been suggested in previous studies using SNP markers and mitochondrial DNA24,38.
The Mixteco Creole cattle show a level of genetic variability similar to that reported in studies of Creole cattle populations in the Americas. In addition, it is more genetically related to other Mexican Creole cattle populations. However, there is evidence of the influence of exotic germplasm, in smaller percentages, from specialized Bos taurus and Bos indicus breeds.
Acknowledgments
The authors wish to express their gratitude to the BIOBOVIS consortium of the CONBIAND Network for providing genotypic information on the reference cattle breeds used in this study.
REFERENCES
1. Fernández de Sierra G, Gómez-Castro AG. Caracterización, utilización y conservación de los recursos zoogenéticos locales. Arch Zootec 2007;56(1):377-378. [ Links ]
2. da Silva A. El plan de acción mundial de la FAO sobre los recursos zoogenéticos y su aplicación en Latinoamérica y el Caribe. RCCA 2014;48(1):35-41. [ Links ]
3. Méndez M, Ávila R, Serrano J, Rosas M, Méndez N. Caracterización morfométrica del bovino criollo mixteco. Arch Zootec 2002;51(194):217-221. [ Links ]
4. Fuentes-Mascorro G, Pérez Vargas E, Carmona Medero MA. Los bovinos criollos de Oaxaca y su importancia. Etnozoología de recursos zoogenéticos Oaxaca y Zulia. México: Editorial Armadillo; 2011:1-9. [ Links ]
5. Parra-Cortés R, Magaña-Magaña MA, Piñeiro-Vázquez A. Intensificación sostenible de la ganadería bovina tropical basada en recursos locales: alternativa de mitigación ambiental para América Latina. ITEA-Inf Tec Econ Agrar 2019;115(4):342-359. [ Links ]
6. FAO. The state of the world’s animal genetic resources for food and agriculture. Italy. Commission on genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations. 2007. [ Links ]
7. Delgado JV, Martínez AM, Acosta A, Álvarez LA, Armstrong E, Camacho E, et al. Genetic characterization of Latin-American creole cattle using microsatellite markers. Anim Genet 2011;43(1):2-10. [ Links ]
8. Martínez AM, Gama LT, Cañón J, Ginja C, Delgado JV, Dunner S, et al. Genetic footprints of Iberian cattle in America 500 years after the arrival of Columbus. PLoS One 2012;7(11):e49066. [ Links ]
9. Gama LT, Martinez AM, Ginja C, Cañon J, Martin-Burriel I, Revidatti MA, et al. Genetic diversity and structure of Iberoamerican livestock breeds. Freitas DA, Lopes da Costa L. Advances in animal health, medicine and production. Cham, Suitzrland: Springer International Publishing; 2020:52-68. [ Links ]
10. Ginja C, Gama LT, Cortes Ó, Delgado JV, Dunner S, García D, et al. Analysis of conservation priorities of Iberoamerican cattle based on autosomal microsatellite markers. Genet Sel 2013;45(1):1-18. [ Links ]
11. Fuentes-Mascorro G, Cruz-Matías J, Pérez-Vargas E, Carmona-Medero MA. Bovinos criollos de Oaxaca. Perezgrovas-Garza RA. Catálogo ilustrado de los bovinos criollos de México. Universidad Autónoma de Chiapas. México 2017:31-45. [ Links ]
12. Hale ML, Burg TM, Steeves TE. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS One 2012;7(9):e45170. [ Links ]
13. FAO. Molecular genetic characterization of animal genetic resources. Italy. Commission on genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations. 2011. [ Links ]
14. FAO. Secondary guidelines for development of national farm animal genetic resources management plans. Measurement of Domestic Animal Diversity (MoDAD). Recommended microsatellite markers. Italy. Commission on genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations. 2004. [ Links ]
15. Yeh F, Boyle T, Ye Z, Xiyan JM, Ye ZH. Popgene Version 1.31 Microsoft window-based freeware for population genetic analysis. BMC Microbiol 1999;12(1):39. [ Links ]
16. Excoffier L. Lischer HEL. An integrated software package for population genetics data analysis. Mol Ecol Resour 2010;10(1):564-567. [ Links ]
17. Ginja C, Gama LT, Cortés O, Burriel IM, Vega-Pla JL, Penedo C, et al. The genetic ancestry of American Creole cattle inferred from uniparental and autosomal genetic markers. Sci Rep 2019;9(1):e11486. [ Links ]
18. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155(2):945-959. [ Links ]
19. FAO. In vivo conservation of animal genetic resources. Italy. Commission on genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations. 2013. [ Links ]
20. Delgado JV, Barba CJ, Aguirre EL, Cabezas R, Cevallos OF, et al. Molecular inference in the colonization of cattle in Ecuador. Res Vet Sci 2020;132:357-368. [ Links ]
21. Ginja C, Penedo MCT, Melucci L, Quiroz J, Martínez-López OR, Revidatti MA, et al. Origins and genetic diversity of New World creole cattle: inferences from mitochondrial and Y chromosome polymorphisms. Anim Genet 2010;41:128-141. [ Links ]
22. Giovambattista G, Ripoli MV, de Luca JC, Mirol PM, Liron JP, Dulout FN. Male-mediated introgression of Bos indicus genes into Argentine and Bolivian creole cattle breeds. Anim Genet 2000;31(5):302-305. [ Links ]
23. Miretti MM, Dunner S, Naves M, Contel EP, Ferro JA. Predominant African-derived mtDNA in Caribbean and Brazilian creole cattle is also found in Spanish cattle (Bos taurus). J Hered 2004;95(5):450-453. [ Links ]
24. Carvajal-Carmona LG, Bermudez N, Olivera-Angel M, et al. Abundant mtDNA diversity and ancestral admixture in Colombian criollo cattle (Bos taurus). Genetics 2003;165(3):1457-1463. [ Links ]
25. Schmidt TL, Jasper ME, Weeks AR, Hoffmann AA. Unbiased population heterozygosity estimates from genome-wide sequence data. Methods Ecol Evol 2021;12:1888-1898. [ Links ]
26. Greenbaum G, Templeton AR, Zarmi Y, Bar-David S. Allelic richness following population founding events--a stochastic modeling framework incorporating gene flow and genetic drift. PLoS One 2014;9(12):e115203. [ Links ]
27. Weir BS. Genetic data analysis. Sunderland, MA: SinauerAssociates. 1990. [ Links ]
28. Caballero A, García-Dorado A. Allelic diversity and its implications for the rate of adaptation. Genetics 2013;195(4):1373-1384. [ Links ]
29. Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 1980;32(3):314-331. [ Links ]
30. Giles BE, Goudet J. Genetic differentiation in Silene dioica metapopulations: estimation of spatiotemporal effects in a successional plant species. Am Nat 1997;149(3):507-526. [ Links ]
31. Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evol 1965;19:395-420. [ Links ]
32. Palacios EY, Álvarez L, Muñoz J. Genetic diversity of Creole hens of the Colombian southwest. Arch Zootec 2016;65(249):73-78. [ Links ]
33. Ocampo RJ, Martínez JF, Martínez R. Assessment of genetic diversity and population structure of Colombian creole cattle using microsatellites. Trop Anim Health Prod 2021;53(1):122. [ Links ]
34. Graffelman J, Weir B. The transitivity of the Hardy-Weinberg law. Forensic Sci Int Genet 2022;58:e102680. [ Links ]
35. Egito AA, Paiva SR, et al. Microsatellite based genetic diversity and relationships among ten Creole and commercial cattle breeds raised in Brazil. BMC Genet 2007;8:83. [ Links ]
36. McTavish EJ, Decker JE, Schnabel RD, Taylor JF, Hillis DM. New world cattle show ancestry from multiple independent domestication events. Proc Natl Acad Sci 2013;110(15):1398-1406. [ Links ]
37. Decker JE, McKay SD, Rolf MM, Kim J, Molina-Alcalá A, Sonstegard TS, et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet 2014;10(3):e1004254. [ Links ]
38. Gautier M, Naves M. Footprints of selection in the ancestral admixture of a New world creole cattle breed. Mol Ecol 2011;20:3128-3143. [ Links ]
Received: September 13, 2022; Accepted: January 09, 2023











 texto em
texto em 


