Introduction
Temperament is an economically relevant trait that impacts animal welfare and traits related to productivity. Bovine temperament is considered to be the most important trait of an animal's personality and comprises a wide range of behaviors, from docility to fear and nervousness or a lack of response, attempts to escape, and aggressive behavior, in which various parameters such as general locomotor activity and reactivity to stress are observable. Temperament is affected by age, experience, sex, handling, maternal effects, environmental factors, genetics, species and breed1,2. To date, several genomic approaches attempted to identify genomic regions and genes in which underlying single nucleotide polymorphisms (SNPs) are associated with temperament, a complex phenotypical trait.
Quantitative trait locus (QTL) mapping uncovered the first evidence of genomic regions associated with behavioral traits in dairy breeds3,4. The detection of QTLs in the genome led to the proposal of candidate genes under the genomic region encompassed by the QTL, which could potentially be responsible for the differences in trait expression. The identification of candidate genes based on their function and possible involvement in bovine temperament has been a strategy for the search for SNPs. Garza-Brenner et al5 selected a group of 19 genes that participate in the dopamine and serotonin pathway, and through a protein-protein interaction (PPI) analysis, they identified four new interacting candidate genes (POMC, NPY, SLC18A2, and FOSFBJ), of which POMC, SLC18A2 and DRD3, HTR2A (selected based on their function) revealed SNPs associated with Exit Velocity (EV) and Pen Score (PS), which are measurements of bovine temperament in a population of Charolais cattle. The same group found that the variations in these genes (DRD3, HTR2A, and POMC) had an effect on bovine growth (birth weight) in a population of Charolais cattle, showing that the identified variations not only had an effect on bovine temperament but also on live weight traits6. Similarly, with the objective of evaluating the potential relationship of two of these SNPs in the DRD3 and HTR2A genes with bovine temperament and growth characteristics, and feed efficiency, a population of Angus, Brangus, and Charolais cattle with temperament assessments was analysed; the results indicated that there was no association with EV and PS, but the SNP in the HTR2A gene was associated with feed efficiency in Brangus cattle7.
Genome-wide association studies (GWAS), based on high-throughput single nucleotide polymorphism (SNP) genotyping technologies, are a relatively recent approach applied to genetic studies of cattle temperament and have allowed the identification of different groups of candidate genes. Lindholm-Perry et al8 analysed a population of the Angus, Hereford, Simmental, Limousin, Charolais, Gelbvieh, and Red Angus breeds to identify genomic regions and genes associated with flight speed (FS); they determined chromosomal regions on BTA 9 and 17 associated and identified within them three genes GRIA2, GLRB, and QKI associated with nearby SNPs. Valente et al9 evaluated a Nellore population using EV to assess their temperament. The NCKAP5, PARK2, DOCK1, ANTXR1, CPE, and GUCY1A2 genes were detected as potential candidates for the trait of interest. Finally, Dos Santos et al10 used a Guzerat population in which reactivity was measured as an indicator of temperament. The genes POU1F1, DRD3, VWA3A, ZBTB20, EPHA6, SNRPF, and NTN4 were proposed as candidate genes responsible for expression of the trait.
In a related context, exome-specific resequencing of specific regions using next-generation sequencing (NGS) technologies has become a powerful technique that allows the identification of SNPs. This method can efficiently capture all variation in the regions of interest. The potential effects can be assessed in an association study, which provides an effective tool to find SNPs affecting a determined trait11. However, due to differences in temperament phenotyping in previous studies, (i.e., each study uses different techniques to assess bovine temperament, pen score, exit velocity, reactivity, which evaluate different aspects of bovine temperament), it is not possible to link information for those genes identified as candidates, or to find a representative biological process, protein-protein interactions between these genes, or a biological path in which these genes converge to visualize how the set of genes explains bovine temperament. Thus, genomic information often remains isolated and needs to be integrated. Hence, the objective was to identify genomic regions and candidate genes associated with temperament in beef cattle through the integration of a GWAS strategy, protein-protein interaction analysis, and SNPs obtained by specific exome resequencing.
Material and methods
Description of animals and biological sample sources
Data and hair samples were obtained from the biobank located at the Animal Biotechnology Laboratory CBG-IPN and were from a cattle population (n= 104) of young Angus (AN, n=63) and Brangus (BR, n=41) bulls, with an average age and body weight of 273 ± 38 d and 272 ± 38 kg, respectively, analysed during two centralized feed efficiency performance tests based on residual feed intake (RFI) in northern Mexico. Data recording and animal management have been previously described by Garza-Brenner et al7. Briefly, animals were fed in a feedlot for a period of 70 d with a pre-trial adaptation period of 20 d, weighed at the beginning and at the end of the test with intervals of 14 d in which the bovine temperament measurements were made.
From the population, a GWAS was performed using a selective genotyping approach following the strategy of the tails of the phenotypic distribution of bovine temperament measured by exit velocity (EV) because it facilitates the detection of phenotypic differences between alleles12. Selective genotyping was achieved by selecting a group of the calmest (n=17; 10-AN and 7-BR) and most temperamental animals (n=17; 9-AN and 8-BR) based on EV values of study population. Temperament was assessed by EV measurements after a stimulus from hair sampling in a chute by measuring the rate of travel over 1.83 m (6 ft) with an infrared sensor (FarmTek Inc., North Wylie, TX, USA). The velocity was calculated as EV= distance (m)/time (s)13,14. It was defined the contrasting temperament groups based on animals’ EV measurements. Animals with EV measurements ≤1.9 m/s were classified as calm, and those with EV scores ≥2.4 m/s were classified as temperamental14,15.
To identify informative SNPs in candidate genes, 91 animals were used. A total of 91 animals were selected as a SNP discovery population: 18 (9 docile; 9 temperamental) of the Angus breed, 68 (44 docile; 24 temperamental) of the Brahman breed, and 5 (2 docile; 3 temperamental) of Charolais breed. From hair samples and ear notches, DNA extraction was performed using the GenElute™ extraction kit (Sigma, St. Louis, Missouri, United States).
GWAS analysis and gene discovery
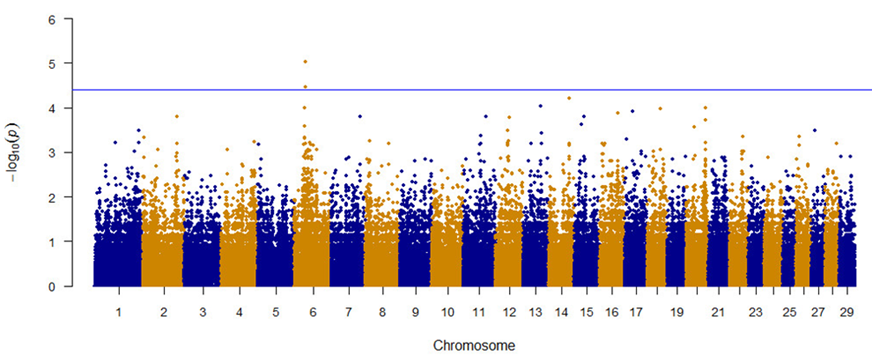
Thirty-four (34) animals were genotyped using the GeneSeek Genomic Profiler HD 150K chip (Neogen, Lincoln, NE). Association analysis and identification of genomic regions associated with bovine temperament were performed with PLINK 1.9 software16. Quality control of the genotypes was performed to identify animals with no assigned genotype or with a low genotyping rate (MIND >0.1). Allele frequency was also evaluated, and those SNPs with lower thresholds (MAF <0.01) were eliminated. Significance threshold was set at P < 3 × 10−5. A Manhattan plot was constructed using qqman: an R package for visualization of GWAS results17. Positions of significant SNPs were identified using the bovine Bos taurus genome (UMD 3.1.1) and Map Viewer software available at the National Center for Biotechnology Information (NCBI). Genes closest to the significant SNPs (within ~350 kb) were also identified with Map Viewer.
Pathway analysis and protein-protein interactions
For the identification of gene pathways, Gene Ontology (GO) term enrichment and protein-protein interaction (PPI) network analysis were performed in the Ensembl genome browser18, Gene Ontology database19, and STRING database20, respectively.
Candidate genes resequencing
With the objective of identifying SNPs in the coding regions and of the SNCA gene and its interacting genes, identified through the protein-protein interaction analysis (PPI), these genes were resequenced in the SNP discovery population. As part of the sequencing strategy, besides the exons, non-coding regions (140 bp before and after each gene-exon) were also analysed. Thus, a customized panel was designed using the Design Studio software (https://designstudio.illumina.com) (Illumina, San Diego, CA, United States) for the AmpliSeq DNA Gene Assay, in which the coding regions and the boundaries of the APP, PARK7, SLC6A2, SNCA, UCHL1, PARK2, SLC18A2, and POMC genes were included, using the Bos taurus UMD 3.1.1 genome as a reference.
DNA quantification was performed in all steps using the Qubit dsDNA HS Assay kit on the Qubit 3.0 fluorometer (Thermo Scientific, Massachusetts, United States). The libraries were prepared using the reference guide for custom panels AmpliSeq (Document # 1000000036408 v04) of Illumina, following the instructions for 2 pools and for 49-96 pairs of primers per pool. The quality and quantification of the libraries were carried out using the Bioanalyzer 2100 equipment (Agilent, California, United States) with the Agilent DNA 1000 kit. Sequencing (paired-end; read length 126 bp) was performed with the MiniSeq ™ Sequencing System.
Bioinformatics analysis of sequencing data
Sequence reads generated by the MiniSeq™ Sequencing System were aligned with the reference genome UMD 3.1.1 of Bos taurus using the Burrows-Wheeler aligner (BWA-MEM) v0.21. The reads were processed using Picard v1.135 (http://broadinstitute.github.io/picard) and cleaned by marking and removing duplicate reads to generate BAM files. Variations were identified using the genomic variant call format (GVCF) workflow with HaplotypeCaller22. SNPs were generated in VCF files and filtered using the following criteria: variant confidence normalized by depth (QD) <2.0, mapping quality (MQ) <40.0, strand bias (FS) >60.0, HaplotypeScore >13.0, MQRankSum <−12.5, and ReadPosRank-Sum <−8.023.
Prediction of the effect of non-coding SNPs on splice sites
To study the effect of the 58 SNPs identified in the non-coding sequences from the exome-specific sequencing of the SNCA and APP genes, the online ESE finder3.0 web interface (http://krainer01.cshl.edu/cgi-bin/tools/ESE3) was used24; the SNCA sequences NC_037333.1 and APP: NC_037328.1 were used as input, introducing them intron by intron (<5000 bp) without and with mutations, according to the location of the SNPs. This process allowed to determine if the SNPs were part of a donor (5´) or acceptor (3´) splice site motif; the programme assigns a score to the input sequence according to the loss of the consensus sequence, so that scores above a default threshold value (donor: 6.67; acceptor: 6.632) are predicted to act as a splice site, allowing the analysis of whether the SNPs affect splice sites motifs.
Results
GWAS analysis and candidate gene identification coupled to protein-protein interaction analysis
Figure 1 depicts a Manhattan plot with the results from the GWAS analysis of SNPs evaluated for their association with temperament in Brangus and Angus cattle. Rs133956611 and rs81144933 were associated with a docile temperament (Table 1). The genes SNCA (alpha-synuclein; GenID 282857) and MMRN1 (multimerin 1; GenID 516574) are located approximately 222.8 and 435.9 Kb upstream respectively, from rs81144933; while the GPRIN3 (GPRIN family member 3; GenID 517995) gene was identified 245.7 Kb downstream of rs133956611.
Table 1 SNPs associated with bovine temperament in Angus and Brangus cattle
| CHR | rs ID | Position pb | Frecuency T D | P-value | |
|---|---|---|---|---|---|
| 6 | rs133956611 | 36,676,986 | 0.14 | 0.67 | 9.2 E-06 |
| 6 | rs81144933 | 36,655,249 | 0.20 | 0.70 | 3.48 E-05 |
T= temperamental; D= docile.
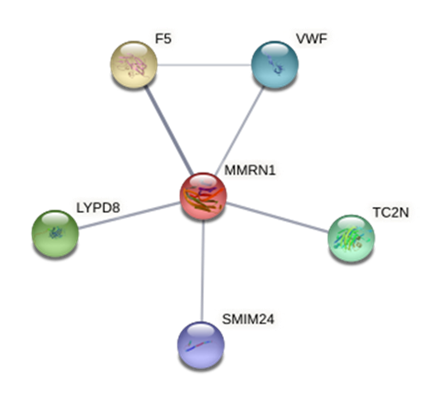
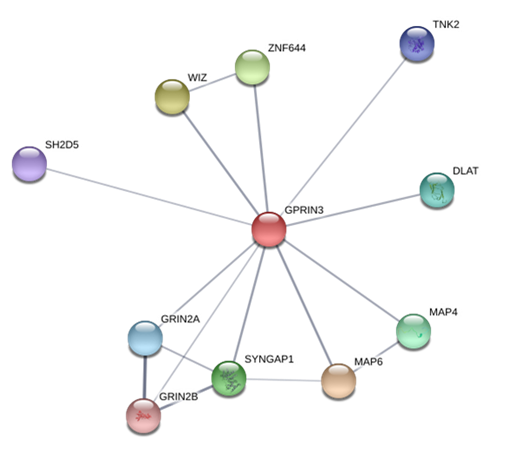
The horizontal line corresponds to a significant threshold of P=3× 10−5 using the identified genes, we proceeded to perform a PPI analysis by querying the STRING20 database. For MMRN1, the PPI analysis showed interactions with genes such as F5 and VWF, involved in the coagulation process (Figure 2), in the Gene Ontology (GO) database, MMRN1 is annotated with the term GO:0007596, named blood coagulation. For GPRIN3, the search engine showed interactions between the phosphorylation process encoded by the LOC790121 and OR6N1 genes with proteins that are mainly involved in cytoskeletal assembly and neurotransmission modulation (Figure 3). The GO database showed that this gene was annotated with the term GO:0031175, biological process named neuron projection development, progression of a neuron projection from its formation to the mature structure.
Finally, SNCA protein, some GO terms identified (GO:0045920, GO:004241 and GO:0014059) were found to be involved in the regulation, synthesis, and secretion of dopamine. Interestingly, the SNCA gene was associated with the terms associated with behavior, including those related to “flight behaviour” and animal responses (through jumping, standing or walking) to internal and external stimuli (terms GO:0007610, GO:0007629 GO:0007628 GO:0007630, respectively).
The PPI analysis indicated that SNCA interacts with APP (β-amyloid precursor protein), PARK7 (parkinsonism associated deglycase), and UCHL1 (ubiquitin C-terminal hydrolase L1) proteins (P= 5.88e-06) involved in adult locomotory behaviour. In addition, the term GO:0008344 reveals strong interactions of SNCA with genes belonging to a neurotransmitter transporter family (SLC6A) in the network (Figure 4).
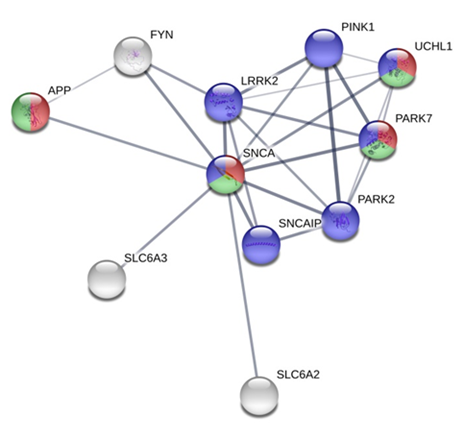
Red nodes annotated with the GO:0008344 term, adult locomotory behavior (p-value 5.88E-06). Green nodes annotated with the GO:0043005 term, neuron projection (p-value 0.000966). Blue nodes annotated with the 05012 KEGG pathway ID, Parkinson s disease (p-value 6.49E-11).
Figure 4 Protein-protein interactions reported for bovine SNCA in the STRING database
Based on their reported functional role, GPRIN3 and, particularly, SNCA genes could be considered as candidate genes associated with cattle temperament, the MMRN1 gene analysis indicate no obvious implications for this trait, however its identification could be important for further analysis.
Genetic variation in candidate genes
According to the PPI analysis results, it was inferred that the APP, PARK7, SLC6A2, UCHL1, PARK2, SLC18A2, and POMC genes were candidates associated with bovine temperament (Table 2). They were resequenced to discover genetic variation to potentially explain cattle temperament. Fifty-eight (58) SNPs were found in the non-coding regions of the SNCA and APP genes. Three SNPs were identified in introns 2 and 3 of the SNCA gene, and 55 SNPs were identified in introns 1, 5, 8, 11, 13, 14, and 17 of the APP gene (Table 3). Fifteen of the 58 SNPs were unique to the Angus breed, 1 in the SNCA gene and the remaining in the APP gene. The remaining SNPs (n= 43) were informative (polymorphic) in the Brahman and Charolais breeds, as opposed to the Angus breed in which they were uninformative (monomorphic). The allelic frequencies and distribution pattern of the SNPs varied according to the breed.
Table 2 Biological functions and processes associated with interacting SNCA genes
| Gene | Description |
|---|---|
| PARK7 | No information in cattle. In humans, protects dopaminergic neurons against oxidative damage and degeneration; indirectly inhibits aggregation of α-synuclein25; thus, mutations in this gene have been demonstrated to cause Parkinson’s disease26. |
| SLC6A2 | No information in cattle. In humans, controls the action of norepinephrine that support arousal, mood, attention, and reactions to stress; thus, it has been associated with temperamental personality dimensions (novelty seeking, harm avoidance, reward dependence, and persistence)27. |
| UCHL1 | No information in cattle. In humans, it is abundantly expressed in neurons and interacts with APP, and SNPs in this gene have been implicated in the neurodegenerative disorders Parkinson’s disease and Alzheimer’s disease28. |
| PARK2 | In cattle, it has been associated with temperament (flight speed)9 and in humans in the functions of dopaminergic neurons due to the mutations in this gene associated with Parkinson’s disease29. |
| SLC18A2 | In cattle, it has been associated with temperament (Pen Score) (Garza-Brenner et al5. It participates in the transport of dopamine, preventing its accumulation and dopaminergic neuron death; therefore, it is a risk factor for Parkinson’s disease30. |
| POMC | In cattle, it has been associated with temperament (Pen Score)5. POMC is the precursor for corticotropic hormone (ACTH), which increases the expression of brain-derived neurotrophic factor (BDNF) responsible for neuron proliferation, differentiation, and survival; thus, it has been implicated in Parkinson’s disease 31. |
Table 3 SNPs identified by specific exome sequencing in each population of APP, PARK7, SLC6A2, SNCA, UCHL1, PARK2, SLC18A2, and POMC genes
| Gene | Position (bp) | Region | Alleles | Frecuency | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Angus | Brahman | Charolais | ||||||||
| Ref | Alt | Ref | Alt | Ref | Alt | Ref | Alt | |||
| SNCA | 36297353 | Intron 3 | G | A | 0.9924 | 0.0076 | 1.0 | 0.0 | ||
| 36297374 | Intron 3 | A | G | 0.8500 | 0.1500 | 1.0 | 0.0 | |||
| 36297422 ¥ | Intron 2 | T | A | 0.5000 | 0.5000 | |||||
| APP | 9674371 | Intron 1 | T | C | 0.9717 | 0.0283 | 1.0 | 0.0 | ||
| 9674423 | Intron 1 | A | C | 0.9403 | 0.0597 | 1.0 | 0.0 | |||
| 9674429 | Intron 1 | T | A | 0.9478 | 0.0522 | 1.0 | 0.0 | |||
| 9674430 ¥ | Intron 1 | T | A | 0.9722 | 0.0278 | |||||
| 9674431* | Intron 1 | A | T | 0.9706 | 0.0294 | 0.9925 | 0.0075 | 1.0 | 0.0 | |
| 9674437 | Intron 1 | T | C | 1.0000 | 0.0 | 0.9000 | 0.1000 | |||
| 9674448 | Intron 1 | T | C | 0.9921 | 0.0079 | 1.0 | 0.0 | |||
| 9674451 | Intron 1 | A | G | 0.9921 | 0.0079 | 0.9000 | 0.1000 | |||
| 9674455* | Intron 1 | G | A | 0.6071 | 0.3929 | 0.0093 | 0.9907 | 0.5000 | 0.5000 | |
| 9770586* | Intron 5 | A | G/T | 0.6944 | 0.3056/0.0 | 0.8772 | 0.0395/0.0833 | 0.8000 | 0.2000/0.0 | |
| 9770593 | Intron 5 | C | T | 0.3507 | 0.6493 | 1.0 | 0.0 | |||
| 9770633 | Intron 5 | G | A | 0.5373 | 0.4627 | 1.0 | 0.0 | |||
| 9803985* | Intron 8 | C | T | 0.9722 | 0.0278 | 0.0944 | 0.9056 | 1.0 | 0.0 | |
| 9803991* | Intron 8 | A | G | 0.9722 | 0.0278 | 0.0909 | 0.9091 | 1.0 | 0.0 | |
| 9806624* | Intron 8 | A | G | 0.9167 | 0.0833 | 0.0574 | 0.9426 | 0.8000 | 0.2000 | |
| 9806672 | Intron 8 | T | C | 0.9769 | 0.0231 | 0.8000 | 0.2000 | |||
| 9806689 | Intron 8 | G | T | 0.9851 | 0.0149 | 1.0 | 0.0 | |||
| 9845631 | Intron 11 | C | A | 1.0000 | 0.0000 | 0.8000 | 0.2000 | |||
| 9845821 | Intron 11 | C | G | 0.7177 | 0.2823 | 1.0 | 0.0 | |||
| 9845862 ¥ | Intron 11 | G | T | 0.8750 | 0.1250 | |||||
| 9845934 | Intron 11 | G | A | 0.9844 | 0.0156 | 1.0 | 0.0 | |||
| 9845944 | Intron 11 | G | A | 0.8750 | 0.1250 | 1.0 | 0.0 | |||
| 9845966 | Intron 11 | G | A | 0.9692 | 0.0308 | 1.0 | 0.0 | |||
| 9845980 | Intron 11 | A | G | 0.8056 | 0.1944 | 1.0 | 0.0 | |||
| 9863873* | Intron 13 | T | C | 0.9722 | 0.0278 | 0.0522 | 0.9478 | 1.0 | 0.0 | |
| 9863960 | Intron 13 | T | C | 0.0818 | 0.9182 | 1.0 | 0.0 | |||
| 9863974¥ | Intron 13 | T | C | 0.6666 | 0.3333 | |||||
| 9863983¥ | Intron 13 | T | C | 0.0 | 1.0 | |||||
| 9863984¥ | Intron 13 | G | T | 0.0 | 1.0 | |||||
| 9866489 | Intron 13 | G | A | 0.8433 | 0.1567 | 1.0 | 0.0 | |||
| 9866528 | Intron 13 | A | G | 0.9925 | 0.0075 | 1.0 | 0.0 | |||
| 9866542¥ | Intron 13 | A | G | 0.9722 | 0.02 | |||||
| 9866545 | Intron 13 | C | T | 0.8433 | 0.1567 | 1.0 | 0.0 | |||
| 9866552¥ | Intron 13 | T | C | 0.9118 | 0.08 | |||||
| 9866569 | Intron 13 | C | T | 0.8624 | 0.1376 | 1.0 | 0.0 | |||
| 9879860 | Intron 13 | T | C | 0.7881 | 0.2119 | 1.0 | 0.0 | |||
| 9880018 | Intron 13 | C | A | 0.5694 | 0.4306 | 1.0 | 0.0 | |||
| 9880025 | Intron 13 | G | T | 0.7787 | 0.2213 | 1.0 | 0.0 | |||
| 9889605¥ | Intron 14 | C | G | 0.6250 | 0.3750 | |||||
| 9889627 | Intron 14 | G | A | 0.9462 | 0.0538 | 1.0 | 0.0 | |||
| 9889677* | Intron 14 | G | T | 0.0 | 1.0 | 0.9925 | 0.0075 | 0.0 | 1.0 | |
| 9889687¥ | Intron 14 | T | C | 0.4167 | 0.5833 | |||||
| 9891054¥ | Intron 14 | C | T | 0.5833 | 0.4167 | |||||
| 9891056¥ | Intron 14 | T | C | 0.5000 | 0.5000 | |||||
| 9891124¥ | Intron 14 | G | T | 0.4000 | 0.6000 | |||||
| 9891130¥ | Intron 14 | A | G | 0.4063 | 0.5938 | |||||
| 9891155¥ | Intron 14 | T | G | 0.4063 | 0.5938 | |||||
| 9918483 | Intron 17 | C | T | 0.9841 | 0.0159 | 1.0 | 0.0 | |||
| 9918506* | Intron 17 | A | G | 0.1389 | 0.8611 | 0.9250 | 0.0750 | 0.2000 | 0.8000 | |
| 9918508 | Intron 17 | C | T | 0.9655 | 0.0345 | 1.0 | 0.0 | |||
| 9918512 | Intron 17 | C | T | 0.9914 | 0.0086 | 1.0 | 0.0 | |||
| 9931517 | Intron 17 | C | G | 0.9924 | 0.0076 | 1.0 | 0.0 | |||
| 9931524 | Intron 17 | C | G | 0.9924 | 0.0076 | 1.0 | 0.0 | |||
| 9931525 | Intron 17 | T | G | 0.9924 | 0.0076 | 1.0 | 0.0 | |||
| 9931529 | Intron 17 | C | T | 1.0000 | 0.0000 | 0.9000 | 0.1000 | |||
* Variations present in the three populations. ¥ Specific variations in the Angus population.
From the 58 SNP´s identified in the non-coding regions of SNCA and APP genes, three SNPs were part of a splice site motif according to established thresholds (donor: 6.67; acceptor: 6.632), as shown in Table 4; the identified SNPs were located in introns 5, 8, and 11. All the splice site motifs were of the acceptor type, that is, they were located at the 3´ end. The SNP g. 9770593 (C/T) did not add or abolish any splice site motif, but only increased the score value, while the SNPs g. 9806689 (G/T) and g. 9845821 (C/G) added and abolished the splice site motifs, respectively.
Table 4 The ESE finder results for non-coding SNPs identified in the SNCA and APP genes
| Gene | Position | Intron | SNP | Site Sequence | Donor/ acceptor | Score | |
|---|---|---|---|---|---|---|---|
| APP | 9770593 | 5 | C | WT | CTCTCCCCTCGTCAGTGCTGTAGTTCAGGT | acceptor | 6.74720 |
| T | M | CTTTCCCCTCGTCAGTGCTGTAGTTCAGGT | acceptor | 7.11480 ↑ | |||
| 9806689 | 8 | G | WT | ------ | ------ | ------ | |
| T | M | CTTTGGATTTGCCAGGCACACTCACCTCCA | acceptor | 6.81380 ↑ | |||
| 9845821 | 11 | C | WT | CTCCTTCCACAACAGAAGGCGCTATTTTAA | acceptor | 6.71530 | |
| G | M | ------ | ------ | ------ | |||
The SNP nucleotide is highlighted in bold in the sequence. WT: wild type. M: sequence with non-coding SNP. ↑ indicates an increased score compared with the wild type sequence.
Discussion
Genomic studies aimed at the exploration of cattle temperament are still scarce, mainly due to the biological complexity of the system, differences in the temperament measurement (objective/subjective), and differences between the studied cattle breeds. In this work, was used the GWAs as an exploratory tool to find candidate genes associated with EV, contrasting by temperament a pool of Angus and Brangus animals. GWAS allowed to identify a genomic region on BTA6 that harbours three candidate genes associated with EV [SNCA (Gen ID 282857), MMRN1 (Gen ID 516574), and GPRIN3 (Gene ID: 517995)]. For these genes, Chen et al32 reported an elevated expression of GPRIN3 in the human brain, and information from UniProtKB33 indicates that the GPRIN3 protein may be involved in neurite outgrowth. However, the literature data (regarding function and interacting genes) strongly supports the bovine SNCA gene as a novel candidate associated with cattle temperament9,34.
The SNCA gene is a highly conserved protein that is abundant in the brain of humans and other species like rats, mice, and monkeys35; it is found in neurons, especially in presynaptic terminals36. The molecular function of SNCA is quite ambiguous, and based on its structure, physical properties, and interacting partners, several hypotheses regarding its normal function in humans have been proposed. For example, it is thought to be involved in the regulation of dopamine release and transport34. Consequently, in humans it plays an important role in neurodegenerative disorders. According to Giasson et al37, aggregates of SNCA protein in humans cause brain lesions that are characteristic of neurodegenerative synucleinopathies. The SNCA gene is associated, in the Kyoto Encyclopedia of Genes and Genomes (KEGG)38, with biological pathways of neurodegenerative diseases such as Alzheimer's disease (ko05010) and Parkinson's disease (ko05012). Both diseases are important brain disorders in humans. Parkinson's disease is characterized by symptoms related to locomotion (involuntary tremor, muscle stiffness, and postural instability), as well as depression and psychosis, and it involves the progressive loss of dopaminergic neurons, with the main feature presenting as the appearance of inclusion bodies called Lewy bodies, the main component of which is SNCA37.
Although the pathological alterations linked to those human diseases cannot be extrapolated to this study model, this biological link provides some evidence to support the findings because the understanding of the relationship between genotype and phenotype in humans was derived from model animals with mutations in orthologous genes. Large animal species, such as dog, pig, sheep and cattle, have been some of the most important model animals, mainly because they are more similar to humans than mice (similar size, genetics, and physiology). Thus, discoveries in humans can serve as a reference to infer effects on bovine temperament39.
Connecting gene networks to explain cattle temperament
Despite scarce attempts to identify genes and genomic regions underlying the genetic architecture of temperament, until now there have been no reports connecting the gene networks associated with this complex trait.
Protein-protein interaction analysis of the SNCA gene allowed to identify and analyse six additional genes, of which two gene members of the SLC family (SLC18A2 and SLC6A4) have already been identified by Garza-Brenner et al5 as interacting genes in a protein-protein network based on dopamine- and serotonin-related genes. These authors also found a SNP located in the SLC18A2 gene that causes a change in the amino acid sequence from alanine to threonine, with significant effects on temperament as measured by pen scores. In addition, the PPI analysis included genes in the PARK family (PARK2 and PARK7), which encode ubiquitin ligase proteins, including parkin RBR E3. The gene PARK2 was identified by Valente et al9 as a candidate gene associated with temperament in Nellore cattle; the authors used EV as a test to evaluate bovine temperament. Multiple studies have used the GWAS strategy to identify genes that are linked to bovine temperament phenotypes8-10, but in none of these cases has it been possible to establish interactions between the identified genes, and the information from each study seems to be isolated and independent, preventing the clarification of the genetic architecture of temperament from the information available to date. In addition, the set of candidate genes does not seem to be associated with a representative biological process that suggests participation in temperament. The identification of SNCA in this work allows to connect the results of Valente et al9 and Garza-Brenner et al5, showing that the genes identified through different strategies (GWAS and protein-protein interaction network analysis) present an important connection. According to these results, it was explored the genetic variation in these genes in cattle with an emphasis on their coding sequences, and the results revealed a high conservation of the exonic sequences in all seven analysed genes. In humans, a low genetic variation has been reported between genes such as SNCA and UCHL140.
Interestingly, and according to previous reports, a high genetic variation was found in the in the non-coding regions of the bovine SNCA and APP genes.
The exact function of the amyloid beta (A4) precursor protein (APP) gene is unknown, but it has been associated with meat softness in pigs41, can participate in the formation of neurons, and is known for its participation in Alzheimer's disease42. Because patients with Alzheimer's disease show the presence and accumulation of both SNCA and APP proteins, it has been proposed that they may be related in some way. Roberts et al43 have shown that SNCA overexpression increases APP levels, and certain mutations in SNCA increase the processing of APP, so the discovery of mutations in the coding regions of these genes could have a functional impact on them and therefore on bovine temperament.
It has been documented that approximately 21% of bovine genes are alternatively spliced44. In silico analyis identified three APP-SNP´s with the potential to have a functional effect in the pre-mRNA splicing process and, therefore, the expression of bovine temperament. As far as known, no different isoforms of the bovine APP gene have been reported, but splice site motifs in bovine genes have been reported to be highly conserved relative to humans44. The human and bovine genes for APP are orthologs, having the same number of amino acids (770) and an identical amino acidic sequence. In humans, 8 different isoforms of the APP gene have been identified due to the alternative splicing in exons 7, 8, and 15, which terminates APP gene expression in neurons, resulting in the implication of a fundamental role in Alzheimer's disease45. Here there was identified 3 SNPs that affect, add, and abolish splice site motifs in the APP gene, in introns 5, 8, and 11, so they could probably affect the final product and have an effect on the expression of bovine temperament.
In the present study, it was used the contrasting phenotype strategy to perform an exploratory GWAS analysis to identify candidate genes for temperament in cattle, and even with the small sample size limitation, the results showing a connection between SNCA and temperament are consistent with larger GWAS studies. Additionally, the coupling of these result with a PPI analysis allowed to establish connections between different genes that were previously identified within the association to the locomotor system. Fine mapping of the candidate genes predicted that the GWAS and PPI genes confirmed the existence of SNPs with the potential to affect bovine temperament. The present study provides valuable information that contributes to the -still scarce- efforts to describe the cattle temperament genetic architecture, and shows that an analytic strategy is appropriate for application in studies with a limited sample size, especially in countries where phenotyping for this complex trait is limited.
Conclusions and implications
A BTA6 genomic region (36,655,249-36,676,986 bp) neighboring the SNCA gene was associated with temperament trait in Angus and Brangus breeds. Six genes, linked to SNCA, were identified as being potentially associated with temperament. From those, the APP gene harboured three SNPs with a potential effect on the pre-mRNA splicing process and expression of bovine temperament.











 text in
text in