Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias pecuarias
versión On-line ISSN 2448-6698versión impresa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.13 no.3 Mérida jul./sep. 2022 Epub 22-Ago-2022
https://doi.org/10.22319/rmcp.v13i3.6067
Articles
Detection of bovine viral diarrhea virus in captive wild artiodactyls in Mexico
a Universidad Nacional Autónoma de México. Facultad de Medicina Veterinaria y Zootecnia, Departamento de Microbiología e Inmunología. Ciudad de México, México.
b Universidad Nacional Autónoma de México. Facultad de Medicina Veterinaria y Zootecnia, Departamento de Patología, Ciudad de México, México.
c Universidad Nacional Autónoma de México. Facultad de Medicina. Departamento de Microbiología y Parasitología. Ciudad de México, México.
d Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria. Centro Nacional de Servicios de Diagnóstico en Salud Animal, Departamento de Validación de Técnicas, Estado de México, México.
Bovine viral diarrhea virus (BVDV) is a pestivirus that infects a broad range of wild and domestic artiodactyls. Pestiviruses can cause a variety of respiratory, gastrointestinal and reproductive disorders that generate substantial losses in the livestock industry. Sharing of water and food sources between wild and domestic populations increases the risk of interspecies pestivirus transmission. Monitoring pestivirus prevalence in both population types is vital. No data is currently available on pestivirus genetic diversity in wild artiodactyl populations in Mexico. Isolation and genetic analysis were done for BVDV from serum samples collected from 371 captive wild artiodactyls in four regions in central and eastern Mexico. Samples from two water buffaloes and one fallow deer were positive for BVDV by RT-PCR. Phylogenetic analysis of the amplified sequences placed them in BVDV subgenotype 1b. A cytopathic strain was isolated from the deer sample. This is the first report of bovine viral diarrhea virus in wild artiodactyls in Mexico and the first to identify the virus subtype.
Key words Pestivirus; BVDV 1b; Genotyping; Isolation; Wild fauna
El virus de la diarrea viral bovina pertenece al género Pestivirus de la familia Flaviviridae. Los pestivirus infectan a un extenso rango de artiodáctilos, silvestres y domésticos, en los cuales ocasionan una gran variedad de desórdenes respiratorios, gastrointestinales y reproductivos que derivan en pérdidas relevantes para la industria pecuaria. El uso compartido de fuentes de agua y alimento entre los ambientes naturales y pecuarios incrementa el contacto directo e indirecto entre animales domésticos y silvestres, lo que aumenta el riesgo de transmisión interespecie de pestivirus. Por este motivo, la vigilancia de enfermedades causadas por pestivirus debería considerar la prevalencia de estos patógenos en animales silvestres. Actualmente se desconoce la diversidad genética de pestivirus en poblaciones silvestres en México. Este grupo de trabajo recolectó muestras de suero de 371 artiodáctilos silvestres en cautiverio en cuatro regiones de cuatro estados de México que incluyen a Veracruz, Querétaro, el Estado de México y la Ciudad de México. Dos muestras de suero de búfalas de agua y una muestra de suero de una gama fueron positivas al virus de la diarrea viral bovina mediante reacción en cadena de la polimerasa con transcripción reversa. El análisis filogenético de las secuencias amplificadas las agrupó dentro del subgenotipo 1b del virus de la diarrea viral bovina. Además, se logró el aislamiento de un virus citopático a partir de la muestra de suero de la gama. Este estudio constituye el primer reporte del virus de la diarrea viral bovina en artiodáctilos silvestres en México.
Palabras clave Pestivirus; VDVB 1b; Genotipificación Aislamiento; Fauna silvestre
Introduction
Bovine viral diarrhea virus (BVDV) belongs to the Pestivirus genus, in the Flaviviridae family. This genus includes bovine viral diarrhea virus 1 (BVDV1) and bovine viral diarrhea virus 2 (BVDV2), among other viruses of veterinary importance. The International Committee on Taxonomy of Viruses (ICTV) classifies BVDV1 as a Pestivirus A species and BVDV2 as a Pestivirus B species1. BVDV1 has 21 subgenotypes (1a-1u), while BVDV2 has 4 subgenotypes (2a-2d)2. Strains of BVDV are classified as cytopathic (CP) or noncytopathic (NCP) biotypes, according to their effect on cultured cells3. The CP biotypes cause vacuolation and cell death, while NCP biotypes cause no visible alterations in cell culture4-5.
The BVDVs infects a wide range of animals belonging to the order Artiodactyla. Until recently, BVDV had been recognized primarily as a pathogenic agent in domestic cattle. It can cause a range of symptoms from subclinical infections to acute infections characterized by inappetence, transient fever, diarrhea, respiratory disorders and reproductive disorders such as abortions, mummifications, congenital defects, stillbirths or the birth of immunotolerant, persistently infected (PI) animals6-7. However, it is now recognized as a causal agent of reproductive, respiratory, immunological and neurological alterations in wild artiodactyls as well8-14.
Vertical transmission of BVDV can occur via transplacental infection, mating or use of infected semen or embryos15. Horizontal transmission occurs through direct or indirect contact with the oral and/or nasal secretions of infected animals12. Factors such as shared land use and animal migration can promote the spread of pestiviruses between domestic and wild animals9. Natural water and food sources are common points of interaction where viruses can spread to a wide variety of hosts12.
BVDV infections generate significant economic losses in the livestock industry in the form of decreased milk production, substandard reproductive performance, growth retardation, congenital defects, predisposition to concomitant diseases and increased mortality of young animals16. Estimated losses are 46 million dollars/year in England, 44.5 million dollars/yr in New Zealand17-18, and 20 million dollars per million births in Denmark19.
Limited data is available on the genetic diversity of circulating pestiviruses in Mexico; indeed, just one study has been done in cattle in which four BVDV genetic variants were identified (subgenotypes 1a, 1b, 1c and, 2a)20. Considering its economic impact in the livestock industry17-19, its ease of transmission between domestic and wild artiodactyls9,12,21, and the presence of BVDV antibodies in white-tailed deer in Mexico, the present study objective was to detect and identify the BVDV genotypes present in wild animals in Mexico to estimate their potential prevalence in the evaluated populations.
Material and methods
Blood samples
A total of 371 blood samples were collected from wild artiodactyls in captivity. The animals were 2 to 3 yr old and included water buffalo (Bubalus bubalis), fallow deer (Dama dama), white-tailed deer (Odocoileus virginianus), eland antelope (Taurotragus oryx) and wild boar (Sus scrofa). They were from Mexico City and the states of Veracruz, Querétaro and Estado de Mexico.
Using Vacutainer TM tubes, from 3 to 6 ml of blood were taken from the jugular vein of each animal. Once the blood coagulated, the samples were centrifuged for 20 min at 2,000 rpm in a clinical centrifuge. The serum was transferred to microcentrifuge tubes and stored at -70 °C until analysis.
RNA extraction
Total RNA was extracted from serum samples using TRIzol TM LS Reagent, following the manufacturer’s instructions. Briefly, 400 µl serum was mixed with 900 µl TRIzol™ LS Reagent by inverting the microfuge tube six times. The mixture was incubated for 5 min at 4 °C, 240 µl chloroform added, homogenized, and incubated for 5 minutes at 4 °C. The mixture was centrifuged for 15 min at 13,000 g and 4 °C. The supernatant (200 µl) was transferred to a nuclease-free tube, 600 µl isopropanol was added and the mixture homogenized. It was incubated at -20 °C for one hour and centrifuged again for 15 min at 13,000 xg and 4 °C. The supernatant was discarded, and the pellet was washed with 1 ml 75% ethanol and centrifuged for 5 min at 13,000 xg and 4 °C. The supernatant was discarded, and the pellet dried for 5 min at room temperature. The pellets were suspended in 20 µl nuclease-free water and stored at -70 °C until use. Total RNA was also extracted from BVDV-free MDBK cells as a negative control and from the NADL reference strain as a positive control in the RT-PCR
Reverse transcription (RT)
Using a 20 µl/reaction total volume, reverse transcription was done with M-MLV Reverse Transcriptase (Thermo-Fisher), following a manufacturer-recommended protocol. Reaction ingredients were 500 ng RNA (1-10 µl), 1 µl random primers (Invitrogen) (0.2 µg/µl), 1 µl dNTP Mix (10 mM each) and nuclease-free water (sufficient to complete 12 µl). These were homogenized by pipetting, incubated for 5 min at 65 °C, and, 4 µl 5X buffer, 2 µl 0.1 M DTT and 1 µl ribonuclease inhibitor (40 U/µl) added. The mixture was homogenized by pipetting and incubated for 2 min at 37 °C. Complementary DNA was synthesized by adding 1 µl M-MLV Reverse Transcriptase (200 U/µl) and incubating for 50 min at 37 °C. The reaction was stopped by heating the mixture to 70 °C for 15 min.
Polymerase chain reaction (PCR) and sequencing
The PCR analysis of the pestivirus 5' UTR region was done using the 5UTRfwd/STAR-Trev, 324/326 and HCV90/HCV368 primers, which amplify 292 bp, 288 bp and 278 bp fragments, respectively22-24. Each reaction was prepared in a 25 µl total volume with 2 µl cDNA, 1 µl sense primer (10 µM), 1 µl antisense primer (10 µM), 1 µl dNTP Mix (10 mM each), 0.2 µl Taq polymerase (5 U/µl), 5 µl buffer (10 X) and 14.8 µl nuclease-free water.
The analysis was run in a thermocycler (Select Cycler II, Select BioProducts) using these parameters: initial denaturation for 4 min at 94 °C; 30 amplification cycles using denaturation for 30 sec at 94 °C, alignment for 30 sec at 56.2 °C (5UTRfwd/STAR-Trev primers), 55 °C (324/326 primers) and 50 °C (HCV90/HCV368 primers), and extension for 30 sec at 72 °C; and final extension for 10 min at 72 °C.
The RT-PCR products were separated by 1% agarose gel electrophoresis, following the methodology described15,25. Agarose gels were stained with GelRed® following the manufacturer’s protocol. The RT-PCR products were viewed with a UV transilluminator and purified with the QIAquick® Gel Extraction Kit (Qiagen GmbH) following the manufacturer's protocol. The amplicons were sequenced in duplicate in both directions with the BigDyeTM Terminator Sequencing Kit on an ABI PRISM® 3130xl Genetic Analyzer at the UNAM Biotechnology Institute.
Phylogenetic analysis
Phylogenetic reconstruction of the 5' UTR region sequences of the detected viruses was done using the MEGA 10 program, with the maximum verisimilitude method and the Kimura 2 substitution method with 1,000 bootstraps26-28.
Viral isolation
Viral isolation was done following standard cell culture techniques29, and using 25 cm2 cell culture dishes with a 70% confluence of MDBK cells (BVDV-free) in MEM supplemented with 5% horse serum. Samples (500 μl) of serum identified as positive by RT-PCR were deposited in the culture dishes and identification of infected MDBK cell cultures done by observing cytopathic effects (vacuolation and lysis) 48 h after incubation, RT-PCR, and sequencing.
The isolation titer was measured using the Reed-Muench method30. Culture supernatant (100 μl) containing the isolated virus and 900 μl MEM supplemented with 5% horse serum were placed in each well of a 96-well microplate. Decimal log dilutions were done in triplicate from 10-1 to 10-8, and the microplates incubated for 48 h at 37 ºC.
Results
The RT-PCR analysis with the 324/326 primers identified BVDV RNA in the serum sample of a two-year-old female fallow deer identified as 7916 (Mexico City). The 5UTRfwd/STAR-Trev and 324/326 primers identified BVDV RNA in serum samples from two six-mo-old water buffaloes identified as BUMA 6 and BUMA 15 (Mil Aguas, Veracruz). Phylogenetic analysis of the 5' UTR amplicons to identify BVDV species and subgenotypes in the sequences of the three positive samples grouped them in BVDV subgenotype 1b (Figure 1).
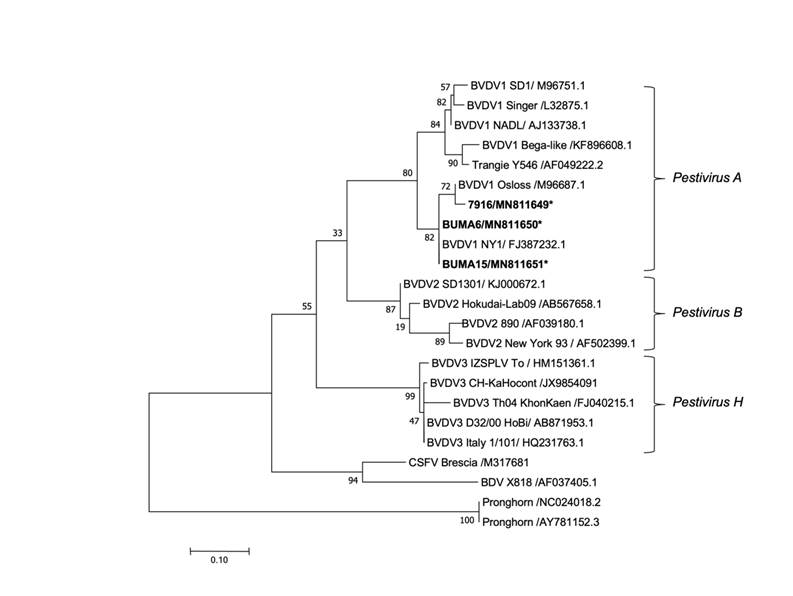
Reconstruction of the phylogenetic tree rooted in the 5' UTR region built with the maximum verisimilitude method using the MEGA 10 program with a Kimura 2 substitution model and 1,000 bootstraps. Reference virus sequences are identified by their GenBank Access numbers, and viral sequences identified in the present study are in bold with an asterisk.
Figure 1 Phylogenetic analysis of isolated pestivirus based on a fragment of the 5' UTR region
Phylogenetic analysis of the 5' UTR region indicated that the BUMA 6 (GenBank #: MN811650) and BUMA 15 (GenBank #: MN811651) sequences exhibited 99.8 % identity with the NY reference strain of BVDV, which belongs to subgenotype 1b. The 7916 (GenBank #: MN811649) sequence exhibited 99.8 % identity with the Osloss reference strain of BVDV, which also belongs to subgenotype 1b.
Use of BVDV-free MDBK cells allowed isolation of a CP biotype virus from the fallow deer (7916) serum; the titer of the isolated BVDV 1b was 106 CCID50/ml. Virus isolation was unsuccessful in the water buffalo (BUMA 6 and BUMA 15) serum samples.
Discussion
The BVDV subgenotype 1b is considered the most widely distributed worldwide in cattle, followed by subgenotypes 1a and 1c2. Subgenotype 1b has been detected in domestic artiodactyls in various countries (including Mexico) on all five continents2,20. In wild animals, subgenotype 1b has been detected in countries such as Canada (bison)31, China (yak)32, Germany (bongo)33 and the United States (alpaca and white-tailed deer)34-35.
To date, only one study has addressed bovine viral diarrhea prevalence in wild animals in Mexico36. Serum samples were collected from white-tailed deer from three states in northeastern Mexico (Coahuila, Nuevo León, and Tamaulipas) and a 63.5 % seroprevalence identified. However, this study only identified antibodies, and did not involve direct identification nor genetic characterization of BVDV. The present study is therefore the first to report on isolation and direct detection of BVDV in wild animals in Mexico, and the first report of BVDV prevalence in wild animals in central and eastern Mexico.
The primers used in the present analyses allow detection of various pestivirus species (Pestivirus A-E, G-H)22-24, but only BVDV1 subgenotype 1b RNA (Pestivirus A) was detected. This was found in approximately 1% of the analyzed wild artiodactyl samples. This proof of BVDV1 subgenotype 1b in wild animals in Mexico suggests that it may transmit between wild and domestic artiodactyl populations. The present results may contribute to design of effective disease control programs that include monitoring of domestic and wild artiodactyls, in addition to vaccination to confer specific protection against the BVDV subgenotypes currently circulating in Mexico.
Infections with the CP or NCP biotypes of BVDV have different implications for disease severity37-38. The NCP biotype is commonly detected in samples associated with respiratory disorders, while the CP biotype is usually detected in samples from animals with reproductive, enteric, or systemic disorders38. Infections with the CP biotype can cause disorders in embryonic development, such as mummifications, hydrocephalus, retinal dysplasia, arthrogryphosis, and abortions39. Intrauterine infections with the NCP biotype that occur between d 42 and 125 of pregnancy can cause persistent infections in the fetus and, consequently, the birth of persistently infected animals that are immunotolerant to BVDV, remain seronegative, and disseminate the virus throughout their lives6,40. Simultaneous infection with the CP and NCP biotypes in PI artiodactyls results in mucosal disease, which is lethal. Therefore, presence of the CP and NCP biotypes in the same farm or location is important because it increases the possibility of co-infections with both biotypes41.
PI bulls can shed between 104 and 107 CCID50/ml BVDV in semen during their lifetime42, a titer similar to the 106 CCID50/ml isolated from a clinically healthy female fallow deer (7916) in the present study. This titer suggests that the doe may have been in an early stage of viremia during an acute, but subclinical, infection which would allow virus detection in the serum sample. It is also similar to the 105.8 DICC50/ml in serum, and 105.8 DICC50/ml to 106.3 DICC50/ml in nasal secretions reported in white-tailed deer12.
Unlike in previous studies, the BVDV CP isolated in the present study was from an animal free of clinical signs. It remains unclear if the viral titer and the absence of clinical signs observed with the isolated CP biotype are related to a recurrent subclinical infection. Of note is that the BVDV isolated from the deer serum was capable of replicating in vitro in bovine cells (MDBK), which suggests that this isolate could infect both species. Further research is required to evaluate its transmission capacity and virulence in both species.
This study is the first report of the detection and isolation of BVDV (Pestivirus A, subgenotype 1b, biotype CP) in water buffalo and fallow deer in Mexico. No other pestivirus species were detected. Further research is needed to better characterize the circulating pestiviruses in wild animal populations in Mexico since they can represent a source of infection for domestic species and vice versa. The present results contribute to understanding the genetic and epidemiological diversity of pestiviruses in the evaluated populations.
Acknowledgements
The research reported here was financed by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica IN217919 “Identificación y caracterización genética de las cepas del virus de la diarrea viral bovina circulantes en poblaciones ganaderas de México”, Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México (FMVZ-UNAM).
REFERENCES
1. Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch Virol 2021;166(9):2633-2648. [ Links ]
2. Yeşilbağ K, Alpay G, Becher P. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses 2017;9(6):E128. [ Links ]
3. Tautz N, Tews BA, Meyers G. The molecular biology of pestiviruses. Adv Virus Res 2015;(93):47-160. [ Links ]
4. Lértora WJ. Diarrea viral bovina: actualización. Rev Vet 2003;14(1):42-51. [ Links ]
5. Rondón I. Diarrea vial bovina: patogénesis e inmunopatología. Rev MVZ Córdoba 2006;11(1):694-704. [ Links ]
6. Strong R, La Rocca SA, Ibata G, Sandvik T. Antigenic and genetic characterisation of border disease viruses isolated from UK cattle. Vet Microbiol 2010;141(3-4):208-215. [ Links ]
7. Brock KV. The many faces of bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract 2004;20(1):1-3. [ Links ]
8. Mattson DE, Baker RJ, Catania JE, Imbur SR, Wellejus KM, Bell RB. Persistent infection with bovine viral diarrhea virus in an alpaca. J Am Vet Med Assoc 2006;228(11):1762-1765. [ Links ]
9. Vilcek S, Nettleton PF. Pestiviruses in wild animals. Vet Microbiol 2006;116(1-3):1-12. [ Links ]
10. Nelson DD, Dark MJ, Bradway DS, Ridpath JF, Call N, Haruna J, et al. Evidence for persistent Bovine viral diarrhea virus infection in a captive mountain goat (Oreamnos americanus). J Vet Diagn Invest 2008;20(6):752-759. [ Links ]
11. Duncan C, Ridpath J, Palmer MV, Driskell E, Spraker T. Histopathologic and immunohistochemical findings in two white-tailed deer fawns persistently infected with Bovine viral diarrhea virus. J Vet Diagn Invest 2008;20(3):289-296. [ Links ]
12. Passler T, Ditchkoff SS, Walz PH. Bovine viral diarrhea virus (BVDV) in white-tailed deer (Odocoileus virginianus). Front Microbiol 2016;(7):945. [ Links ]
13. Wolff PL, Schroeder C, McAdoo C, Cox M, Nelson DD, Evermann JF, et al. Evidence of bovine viral diarrhea virus infection in three species of sympatric wild ungulates in Nevada: life history strategies may maintain endemic infections in wild populations. Front Microbiol 2016;(7):292. [ Links ]
14. Salgado R, Hidalgo-Hermoso E, Pizarro-Lucero J. Detection of persistent pestivirus infection in pudú (Pudu puda) in a captive population of artiodactyls in Chile. BMC Vet Res 2018;14(1):37. [ Links ]
15. Houe H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol 1999;64(2-3):89-107. [ Links ]
16. Houe H. Economic impact of BVDV infection in dairies. Biologicals 2003;31(2):137-143. [ Links ]
17. Bennett RM, Christiansen K, Clifton-Hadley RS. Estimating the costs associated with endemic diseases of dairy cattle. J Dairy Res 1999;66(3):455-459. [ Links ]
18. Heuer C, Healy A, Zerbini C. Economic effects of exposure to bovine viral diarrhea virus on dairy herds in New Zealand. J Dairy Sci 2007;90(12):5428-5438. [ Links ]
19. Houe H, Pedersen KM, Meyling A. A computarized spread sheet model for calculating total annual national losses due to bovine viral diarrhoea virus infection in dairy herds and sensitivity analysis of selected parameters. In: Proc Second Symp Pestiviruses. Annecy, France. 1993:179-184. [ Links ]
20. Gómez-Romero N, Basurto-Alcántara FJ, Verdugo-Rodríguez A, Bauermann FV, Ridpath JF. Genetic diversity of bovine viral diarrhea virus in cattle from Mexico. J Vet Diagn Invest 2017;29(3):362-365. [ Links ]
21. Kirkland PD, Frost MJ, King KR, Finlaison DS, Hornitzky CL, Gu X, et al. Genetic and antigenic characterization of Bungowannah virus, a novel pestivirus. Vet Microbiol 2015;178(3-4):252-259. [ Links ]
22. Mahony TJ, McCarthy FM, Gravel JL, Corney B, Young PL, Vilcek S. Genetic analysis of bovine viral diarrhoea viruses from Australia. Vet Microbiol 2005;106(1-2):1-6. [ Links ]
23. Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol 1994;136(3-4):309-323. [ Links ]
24. Ridpath JF, Lovell G, Neill JD, Hairgrove TB, Velayudhan B, Mock R. Change in predominance of Bovine viral diarrhea virus subgenotypes among samples submitted to a diagnostic laboratory over a 20-year time span. J Vet Diagn Invest 2011;23(2):185-193. [ Links ]
25. Armstrong JA, Schulz JR. Agarose gel electrophoresis. Curr Protoc Essen Lab Tech 2015;10(1):7.2.1-7.2.22. [ Links ]
26. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16(2):111-120. [ Links ]
27. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39(4):783-791. [ Links ]
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35(6):1547-1549. [ Links ]
29. Warner DR, Sakai D, Sandell LL. Mammalian cell culture. Curr Protoc Essen Lab Tech 2015;10(1):4.3.1-4.3.33. [ Links ]
30. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 1938;27(3):493-497. [ Links ]
31. Deregt D, Tessaro SV, Baxi MK, Berezowski J, Ellis JA, Wu JTY, et al. Isolation of bovine viral diarrhoea viruses from bison. Vet Rec 2005;157(15):448-450. [ Links ]
32. Gong X, Liu L, Zheng F, Chen Q, Li Z, Cao X, et al. Molecular investigation of bovine viral diarrhea virus infection in yaks (Bos gruniens) from Qinghai, China. Virol J 2014;(11):29. [ Links ]
33. Becher P, Orlich M, Kosmidou A, König M, Baroth M, Thiel HJ. Genetic diversity of pestiviruses: identification of novel groups and implications for classification. Virology 1999;262(1):64-71. [ Links ]
34. Passler T, Walz PH, Ditchkoff SS, Brock KV, Deyoung RW, Foley AM, et al. Cohabitation of pregnant white-tailed deer and cattle persistently infected with Bovine viral diarrhea virus results in persistently infected fawns. Vet Microbiol 2009;134(3-4):362-367. [ Links ]
35. Goyal SM, Bouljihad M, Haugerud S, Ridpath JF. Isolation of bovine viral diarrhea virus from an alpaca. J Vet Diagn Invest 2002;14(6):523-525. [ Links ]
36. Cantu A, Ortega-S JA, Mosqueda J, Garcia-Vazquez Z, Henke SE, George JE. Prevalence of infectious agents in free-ranging white-tailed deer in northeastern Mexico. J Wildl Dis 2008;44(4):1002-1007. [ Links ]
37. Fulton RW, Ridpath JF, Saliki JT, Briggs RE, Confer AW, Burge LJ, et al. Bovine viral diarrhea virus (BVDV) 1b: predominant BVDV subtype in calves with respiratory disease. Can J Vet Res 2002;66(3):181-190. [ Links ]
38. Odeón AC, Risatti G, Kaiser GG, Leunda MR, Odriozola E, Campero CM, et al. Bovine viral diarrhea virus genomic associations in mucosal disease, enteritis and generalized dermatitis outbreaks in Argentina. Vet Microbiol 2003;96(2):133-144. [ Links ]
39. Brownlie J. The pathogenesis of bovine virus diarrhoea virus infections. Rev Sci Tech 1990;9(1):43-59. [ Links ]
40. Falkenberg SM, Dassanayake RP, Walz P, Casas E, Neill JD, Ridpath JF. Frequency of bovine viral diarrhea virus detected in subpopulations of peripheral blood mononuclear cells in persistently infected animals and health outcome. Vet Immunol Immunop 2019;(207):46-52. [ Links ]
41. Rondena M, Lorenzi V, Binanti D, Gelmetti D, Pravettoni D, Finazzi M, et al. Immune-depletion related to bovine viral diarrhoea virus in a heifer with naturally occurring mucosal disease. J Comp Pathol 2009;141(4):306. [ Links ]
42. Givens MD, Riddell KP, Edmondson MA, Walz PH, Gard JA, Zhang Y, et al. Epidemiology of prolonged testicular infections with bovine viral diarrhea virus. Vet Microbiol 2009;139(1-2):42-51. [ Links ]
Received: September 20, 2021; Accepted: December 21, 2021











 texto en
texto en 


